
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,030,665 – 9,030,817 |
| Length | 152 |
| Max. P | 0.557723 |

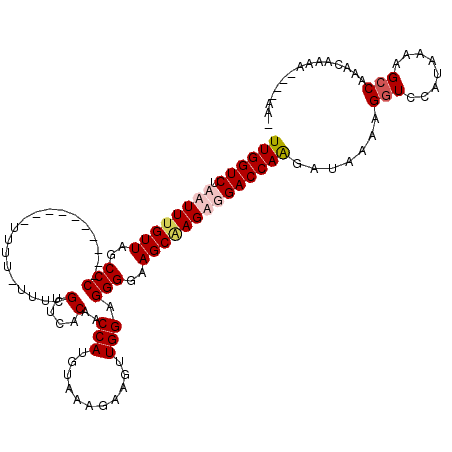
| Location | 9,030,665 – 9,030,783 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.02 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -15.60 |
| Energy contribution | -16.73 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9030665 118 - 22224390 UUGGUCUAAUUUGUUAGCCCAUUUUUUUUUUUUU-UUUCGUUCACAACCAUGUAAAGAAGUUGGAGGGGAAGCAAGUUGACCAGGAUAAAAGGUCCAUUAAAGCCAAAGAAAAAACAAA- .(((((.((((((((..(((.((((..(((((((-...(((........))).)))))))..))))))).)))))))))))))((((.....)))).......................- ( -26.40) >DroSec_CAF1 18466 104 - 1 UUGGUCUAAUUUGUUAGCCC----------UUUU-UUUCGUUCACAACCAUGUAAAGAAGUUGGAGGGGAAGCAAGUUGACCAGGAUAAAAGGUCCAUUAAAGCCAAGCAAAA----AA- .(((((.((((((((..(((----------(((.-((((....(((....)))...))))..))))))..)))))))))))))((((.....)))).................----..- ( -28.40) >DroEre_CAF1 18674 110 - 1 UUGGUCUAAUUCGUUAGCCC----------UUUUAAUUUGUUCACAACCAUGUAAAGAAGUUGGAGGGGAAGCAAGAGGACCAAGAUAAAAGGUCCAUAAAAGCCAAAGAAAAUACAAGU (((((.......(((..(((----------(((((((((.((.(((....))).)).)))))))))))).)))....(((((.........)))))......)))))............. ( -29.80) >DroYak_CAF1 19036 103 - 1 UUGGUCUAAUUUGUUAGCCC----------UUUUUGUUCGUUCACAACCAUGUAAAGAAGUUGGAGGGGAAGCGAGAGGACCAAGAUAAAAGAUCCUUAAAAGCCAAACGAAA------- (((((((..((((((..(((----------(((...(((....(((....)))...)))...))))))..)))))).))))))).............................------- ( -21.80) >consensus UUGGUCUAAUUUGUUAGCCC__________UUUU_UUUCGUUCACAACCAUGUAAAGAAGUUGGAGGGGAAGCAAGAGGACCAAGAUAAAAGGUCCAUAAAAGCCAAACAAAA____AA_ ((((((.((((((((..(((...................(....)..(((...........))).)))..)))))))))))))).......(((........)))............... (-15.60 = -16.73 + 1.12)


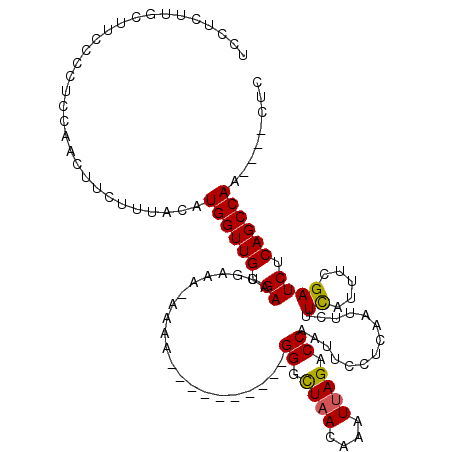
| Location | 9,030,704 – 9,030,817 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.71 |
| Mean single sequence MFE | -15.89 |
| Consensus MFE | -10.90 |
| Energy contribution | -10.78 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9030704 113 + 22224390 UCAACUUGCUUCCCCUCCAACUUCUUUACAUGGUUGUGAACGAAA-AAAAAAAAAAAAAUGGGCUAACAAAUUAGACCAAUUCCUCAAUUCUUCAUUUCGAUCUCAGCCAA----CUC ..............................((((((.((.(((((-.............(((.((((....)))).)))................))))).)).)))))).----... ( -16.65) >DroSec_CAF1 18501 103 + 1 UCAACUUGCUUCCCCUCCAACUUCUUUACAUGGUUGUGAACGAAA-AAAA----------GGGCUAACAAAUUAGACCAAUUCCUCAAUUCUUCAUUUCGAUCUCAGCCAA----CUC ..............................((((((.((.(((((-..((----------(((((((....)))).))((((....)))))))..))))).)).)))))).----... ( -17.60) >DroEre_CAF1 18714 104 + 1 UCCUCUUGCUUCCCCUCCAACUUCUUUACAUGGUUGUGAACAAAUUAAAA----------GGGCUAACGAAUUAGACCAAUUUCUGAAUUCUUCAUUUCGAUCUCAGCCAA----CUC ..............................((((((.((.(.........----------((.((((....)))).)).......(((........)))).)).)))))).----... ( -14.30) >DroYak_CAF1 19069 104 + 1 UCCUCUCGCUUCCCCUCCAACUUCUUUACAUGGUUGUGAACGAACAAAAA----------GGGCUAACAAAUUAGACCAAUUCCCAAAUUCUUCACUUCGAUCUCAGCCAA----CUC ..............................((((((.((.(((((.....----------)((((((....)))).))..................)))).)).)))))).----... ( -15.10) >DroAna_CAF1 35006 105 + 1 UCCUACUUCUUCCCCUCCAACUUCUUUACAUGGUUGUGAACAAA--AAUG----------GGAUUUAAAAAUUAGACCAAUUCCUCUC-GAUUUGUUUCGAUCUCAGCCAGCAGAUUC ......................(((.....((((((.((.....--...(----------(((((.............))))))..((-((......)))))).))))))..)))... ( -15.82) >consensus UCCUCUUGCUUCCCCUCCAACUUCUUUACAUGGUUGUGAACGAAA_AAAA__________GGGCUAACAAAUUAGACCAAUUCCUCAAUUCUUCAUUUCGAUCUCAGCCAA____CUC ..............................((((((.((.....................((.((((....)))).))..............((.....)))).))))))........ (-10.90 = -10.78 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:06 2006