
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,001,059 – 9,001,163 |
| Length | 104 |
| Max. P | 0.688402 |

| Location | 9,001,059 – 9,001,163 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 92.71 |
| Mean single sequence MFE | -24.73 |
| Consensus MFE | -17.34 |
| Energy contribution | -16.90 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
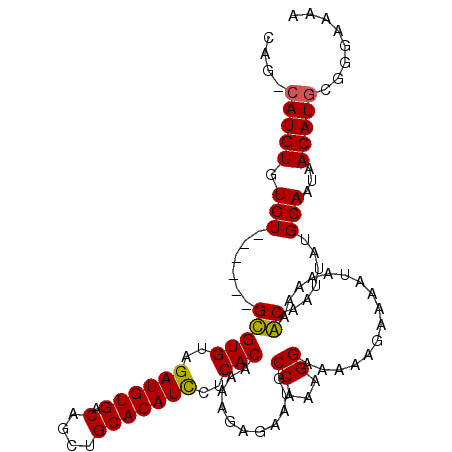
>X_DroMel_CAF1 9001059 104 + 22224390 CAG-CAUGUGUGU------GCGUGUAGAUGUGACAGCUGCACAUCCUCACAAAAGAGAAUGCCAAAGGAAAAAGAAAAUAAAAAGCAAUUAUGCAAUAACAUACGGGAAAA ...-..(((((((------..(((..((((((.(....)))))))..)))..................................(((....)))....)))))))...... ( -21.10) >DroSec_CAF1 1689 104 + 1 CAG-CAUGUGUGU------GCGUGUAGAUGUGACAGCUGCACAUUCUCACAAAAGAGAAUGCCAAAGGAAAAAGAAAAUAAAAAGCAAUUAUGCAAUAACAUGCGGGAAAA ..(-(((((...(------((((((((.((...)).))))))((((((......))))))................................)))...))))))....... ( -26.90) >DroEre_CAF1 1829 103 + 1 CAG-CAUGUGUGU------GUGUGUAGAUGUGACAGCUGCACAUCCUCACAAAAGAGAAUGCCAAAGGAAAAAGAAAAUAA-AAACAAUUAUGCAAUAACAUGCGGGAAAA ..(-(((((.(((------(((((..((((((.(....)))))))..)))...........((...)).............-........)))))...))))))....... ( -23.20) >DroYak_CAF1 1759 111 + 1 CAACCAUGUGUGUGUGUGCGUGUGUAGAUGUGACAGCUGCACAUCCUCACAAAAGAGAAUGCCGAAGGAAAAGGAAAAUAAAAAACAAUUAUGCAAUAACAUGCGGGAAAA ...((.(((((((...((((((((..((((((.(....)))))))..)))...........((.........))................)))))...))))))))).... ( -27.70) >consensus CAG_CAUGUGUGU______GCGUGUAGAUGUGACAGCUGCACAUCCUCACAAAAGAGAAUGCCAAAGGAAAAAGAAAAUAAAAAACAAUUAUGCAAUAACAUGCGGGAAAA ....(((((.(((......(((((..((((((.(....)))))))..)))...........((...))................))......)))...)))))........ (-17.34 = -16.90 + -0.44)


| Location | 9,001,059 – 9,001,163 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 92.71 |
| Mean single sequence MFE | -21.71 |
| Consensus MFE | -16.27 |
| Energy contribution | -15.65 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
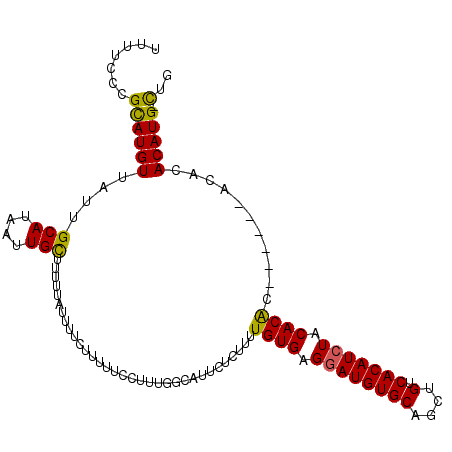
>X_DroMel_CAF1 9001059 104 - 22224390 UUUUCCCGUAUGUUAUUGCAUAAUUGCUUUUUAUUUUCUUUUUCCUUUGGCAUUCUCUUUUGUGAGGAUGUGCAGCUGUCACAUCUACACGC------ACACACAUG-CUG .......((((((...(((.....((((....................)))).........(((.((((((((....).))))))).)))))------)...)))))-).. ( -24.05) >DroSec_CAF1 1689 104 - 1 UUUUCCCGCAUGUUAUUGCAUAAUUGCUUUUUAUUUUCUUUUUCCUUUGGCAUUCUCUUUUGUGAGAAUGUGCAGCUGUCACAUCUACACGC------ACACACAUG-CUG .......((((((...((((((..((((....................))))(((((......)))))))))))(((((.......))).))------....)))))-).. ( -22.45) >DroEre_CAF1 1829 103 - 1 UUUUCCCGCAUGUUAUUGCAUAAUUGUUU-UUAUUUUCUUUUUCCUUUGGCAUUCUCUUUUGUGAGGAUGUGCAGCUGUCACAUCUACACAC------ACACACAUG-CUG .......(((......)))..........-..................(((((.......((((.((((((((....).))))))).)))).------......)))-)). ( -21.14) >DroYak_CAF1 1759 111 - 1 UUUUCCCGCAUGUUAUUGCAUAAUUGUUUUUUAUUUUCCUUUUCCUUCGGCAUUCUCUUUUGUGAGGAUGUGCAGCUGUCACAUCUACACACGCACACACACACAUGGUUG ........(((((...(((.........................(....)..........((((.((((((((....).))))))).)))).))).......))))).... ( -19.20) >consensus UUUUCCCGCAUGUUAUUGCAUAAUUGCUUUUUAUUUUCUUUUUCCUUUGGCAUUCUCUUUUGUGAGGAUGUGCAGCUGUCACAUCUACACAC______ACACACAUG_CUG .......((((((....(((....))).................................((((.((((((((....).))))))).))))...........))))).).. (-16.27 = -15.65 + -0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:01 2006