
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,988,786 – 8,988,897 |
| Length | 111 |
| Max. P | 0.937854 |

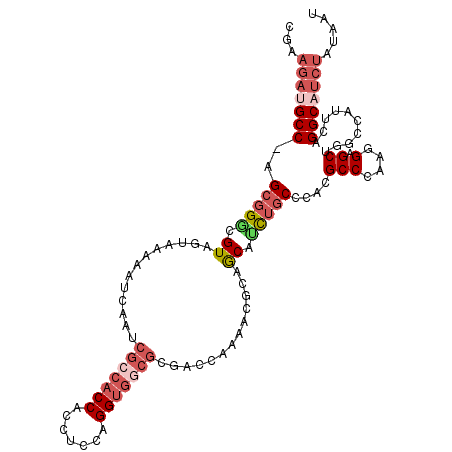
| Location | 8,988,786 – 8,988,897 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.88 |
| Mean single sequence MFE | -39.97 |
| Consensus MFE | -22.50 |
| Energy contribution | -24.05 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8988786 111 + 22224390 GGAAGAUGCC-GGCGGGCGUAGUAAAAAUCAAUCGCCACCACCUGCAGGUGGCGUGAACAAAACGCAGCAUUUGCCCACGCCCAAGGAGCUGGCCAUUCAGGCAUCUAAAAU ...(((((((-...((((((.(((((........((((((.......))))))(((.......)))....)))))..))))))..((......)).....)))))))..... ( -41.70) >DroPse_CAF1 657 102 + 1 -------GCCGAGUGGCCGUGGGGCCAAUCGUUCGCCACCGGCGCCUGGUAAAAUGCCCAAA---CAACAGCUUCCCACGCCCAAAGAGCUGUCCAUGCAGGCCUCGAUGAU -------((((.(((((....((((.....)))))))))))))(((((......((......---))(((((((............))))))).....)))))......... ( -31.40) >DroSec_CAF1 705 111 + 1 CGAAGAUGCC-GGCCGGCGUAGUAAAAACCAAUCGCCACCACCUCCAGGUGGCGCGACCAAAACGCAACAUUUGCCCACGCCCAAGGAGCUGGCCAUUCAGGCAUCUAUAAU ...(((((((-(((((((...(((((........((((((.......))))))(((.......)))....)))))((........)).))))))).....)))))))..... ( -42.90) >DroSim_CAF1 2449 111 + 1 CGAAGAUGCC-GGCGGGCGUAGUAAAAACCAAUCGCCACCACCUCCAGGUGGCGCGACCAAAACGCAGCAUUUGCCCACGCCCAAGGAGCUGGCCAUUCAGGCAUCUAUAAU ...(((((((-...((((((.(((((........((((((.......))))))(((.......)))....)))))..))))))..((......)).....)))))))..... ( -43.60) >DroEre_CAF1 717 111 + 1 CGAAGAUGCC-AGCGGACGUGGCAAAAAUCGAUCGCCACCACCUCCAGGUGGCGCGACCAAGCCCCAGCACCUGCCCACGCCCAAGGAGCUCUCUAUUCAGGCAUCUAUAAU ...(((((((-...((.(((((((....(((..(((((((.......))))))))))....((....))...)).))))).))..(((....))).....)))))))..... ( -40.50) >DroYak_CAF1 2330 111 + 1 CGAAGAUGCC-AGCGGGCGUGGCAAAAAUCGAUCGCCACCACCUCCCGGGGCCGUGAACAAAACCCAGCACCUGCCCACGCCCAAGGAGCUGUCUAUCCAGGCCUCUUUAAU .(((((.(((-...((((((((........(.((((..((.((....))))..)))).)........((....))))))))))..(((........))).))).)))))... ( -39.70) >consensus CGAAGAUGCC_AGCGGGCGUAGUAAAAAUCAAUCGCCACCACCUCCAGGUGGCGCGACCAAAACGCAGCAUCUGCCCACGCCCAAGGAGCUGGCCAUUCAGGCAUCUAUAAU ...(((((((..(((((.((.............(((((((.......))))))).............)).)))))....(((....).))..........)))))))..... (-22.50 = -24.05 + 1.56)


| Location | 8,988,786 – 8,988,897 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 79.88 |
| Mean single sequence MFE | -47.52 |
| Consensus MFE | -32.28 |
| Energy contribution | -32.65 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.34 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8988786 111 - 22224390 AUUUUAGAUGCCUGAAUGGCCAGCUCCUUGGGCGUGGGCAAAUGCUGCGUUUUGUUCACGCCACCUGCAGGUGGUGGCGAUUGAUUUUUACUACGCCCGCC-GGCAUCUUCC .....(((((((......(((.((......((((((((((((........))))))))))))....)).)))(((((((..............)).)))))-)))))))... ( -43.84) >DroPse_CAF1 657 102 - 1 AUCAUCGAGGCCUGCAUGGACAGCUCUUUGGGCGUGGGAAGCUGUUG---UUUGGGCAUUUUACCAGGCGCCGGUGGCGAACGAUUGGCCCCACGGCCACUCGGC------- ....(((..(((.((...((((((((((.......))).)))))))(---(((((........)))))))).)))..))).(((.(((((....))))).)))..------- ( -41.20) >DroSec_CAF1 705 111 - 1 AUUAUAGAUGCCUGAAUGGCCAGCUCCUUGGGCGUGGGCAAAUGUUGCGUUUUGGUCGCGCCACCUGGAGGUGGUGGCGAUUGGUUUUUACUACGCCGGCC-GGCAUCUUCG .....(((((((.....((((..(.....)((((((((((.....)))...(..(((((((((((....)))))).)))))..)......)))))))))))-)))))))... ( -50.60) >DroSim_CAF1 2449 111 - 1 AUUAUAGAUGCCUGAAUGGCCAGCUCCUUGGGCGUGGGCAAAUGCUGCGUUUUGGUCGCGCCACCUGGAGGUGGUGGCGAUUGGUUUUUACUACGCCCGCC-GGCAUCUUCG .....(((((((......((((.((((...(((((((.((((........)))).)))))))....)))).))))((((..((((....))))....))))-)))))))... ( -50.00) >DroEre_CAF1 717 111 - 1 AUUAUAGAUGCCUGAAUAGAGAGCUCCUUGGGCGUGGGCAGGUGCUGGGGCUUGGUCGCGCCACCUGGAGGUGGUGGCGAUCGAUUUUUGCCACGUCCGCU-GGCAUCUUCG .....(((((((......(((.....)))((((((((.((((.((....))((((((((((((((....)))))).))))))))..))))))))))))...-)))))))... ( -50.00) >DroYak_CAF1 2330 111 - 1 AUUAAAGAGGCCUGGAUAGACAGCUCCUUGGGCGUGGGCAGGUGCUGGGUUUUGUUCACGGCCCCGGGAGGUGGUGGCGAUCGAUUUUUGCCACGCCCGCU-GGCAUCUUCG ....((((.((((((.....((.(((((.(((((((((((((........))))))))).)))).))))).))(((((((.......)))))))..)))..-))).)))).. ( -49.50) >consensus AUUAUAGAUGCCUGAAUGGACAGCUCCUUGGGCGUGGGCAAAUGCUGCGUUUUGGUCACGCCACCUGGAGGUGGUGGCGAUCGAUUUUUACCACGCCCGCC_GGCAUCUUCG .....(((((((.....((....))...(((((((((.((((........))))....(((((((.......)))))))...........)))))))))...)))))))... (-32.28 = -32.65 + 0.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:42 2006