| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,981,905 – 8,982,019 |
| Length | 114 |
| Max. P | 0.979413 |

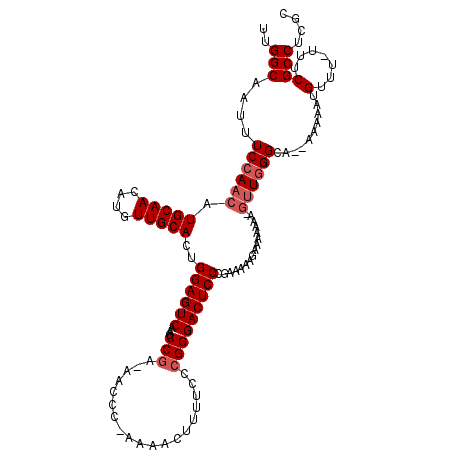
| Location | 8,981,905 – 8,982,019 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.74 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -28.67 |
| Energy contribution | -28.63 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8981905 114 + 22224390 GCGAGGCGAAA-AAACAUUUUU--UGCCCAAC-UUUUUUUCUUUUUCGGGAGUCCCGGGAAAAGUUUU-GGGUU-UCGGCUUGACUCCAGUGCAACAUGUUGCAUGUUGGAAAUUGCCAA ..(.(((((((-(.....))))--)))))(((-((.....(((((((.((....)).)))))))....-)))))-..(((.....((((((((((....))))))..))))....))).. ( -35.50) >DroSec_CAF1 10441 114 + 1 GCGAGGCGAAA-AAACAUUUUU--UGCCCAAC-UUUUUUUCUUUUUCGGGAGUCCCGGGAAAAGUUUU-GGGUU-UCGGCUUGACUCCAGUGCAACAUGUUGCAUGUUGGAAAUUGCCAA ..(.(((((((-(.....))))--)))))(((-((.....(((((((.((....)).)))))))....-)))))-..(((.....((((((((((....))))))..))))....))).. ( -35.50) >DroSim_CAF1 6180 114 + 1 GCGAGGCGAAA-AAACAUUUUU--UGCCCAAC-UUUUUUUCUUUUUCGGGAGUCCCGGGAAAAGUUUU-GGGUU-UCGGCUUGACUCCAGUGCAACAUGUUGCAUGUUGGAUAUUGCCAA ..(.(((((((-(.....))))--)))))(((-((.....(((((((.((....)).)))))))....-)))))-..(((.....((((((((((....))))))..))))....))).. ( -35.50) >DroEre_CAF1 4091 117 + 1 GCCAGGCGAAAAAAACAUUUUUUUUCCCCAAC-UUUUUUCUUAUUUCGGGAGUCCCGGGAAAAGUUUU-GGGUU-UCGGCUUGACUCCAGUGCAACAUGUUGCAUGUUGGAAAUUGCCAA ((((((((((((((.....)))))))(((((.-.(((((((.....((((...)))))))))))..))-)))..-...)))))..((((((((((....))))))..))))....))... ( -35.70) >DroYak_CAF1 3106 119 + 1 GCGAGGCGAAA-AAACAUUUUUUUUUCCAAUUUUUUUUUCUUAUUUUGGGAGUCCCGGGAAAAGUUUUUGGGUUUUGGGUUUGACUCCAGUGCAACAUGUUGCAUGUUGGAAAUUGCCAA ....(((((..-...........((((((((.................((((((((((((...(....)...)))))))....))))).((((((....))))))))))))))))))).. ( -30.71) >consensus GCGAGGCGAAA_AAACAUUUUU__UGCCCAAC_UUUUUUUCUUUUUCGGGAGUCCCGGGAAAAGUUUU_GGGUU_UCGGCUUGACUCCAGUGCAACAUGUUGCAUGUUGGAAAUUGCCAA ....(((((......((........((((.....(((((((.....((((...))))))))))).....))))........))..((((((((((....))))))..))))..))))).. (-28.67 = -28.63 + -0.04)

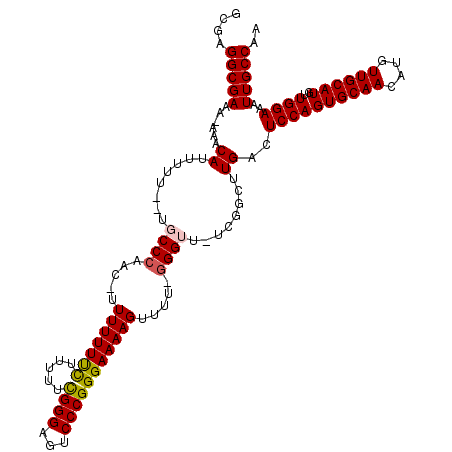
| Location | 8,981,905 – 8,982,019 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.74 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -21.03 |
| Energy contribution | -21.83 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8981905 114 - 22224390 UUGGCAAUUUCCAACAUGCAACAUGUUGCACUGGAGUCAAGCCGA-AACCC-AAAACUUUUCCCGGGACUCCCGAAAAAGAAAAAAA-GUUGGGCA--AAAAAUGUUU-UUUCGCCUCGC (((((...(((((...(((((....))))).)))))....)))))-.....-....(((((..((((...)))).))))).......-((.(((((--((((....))-))).)))).)) ( -29.30) >DroSec_CAF1 10441 114 - 1 UUGGCAAUUUCCAACAUGCAACAUGUUGCACUGGAGUCAAGCCGA-AACCC-AAAACUUUUCCCGGGACUCCCGAAAAAGAAAAAAA-GUUGGGCA--AAAAAUGUUU-UUUCGCCUCGC (((((...(((((...(((((....))))).)))))....)))))-.....-....(((((..((((...)))).))))).......-((.(((((--((((....))-))).)))).)) ( -29.30) >DroSim_CAF1 6180 114 - 1 UUGGCAAUAUCCAACAUGCAACAUGUUGCACUGGAGUCAAGCCGA-AACCC-AAAACUUUUCCCGGGACUCCCGAAAAAGAAAAAAA-GUUGGGCA--AAAAAUGUUU-UUUCGCCUCGC (((((....((((...(((((....))))).)))).....)))))-.....-....(((((..((((...)))).))))).......-((.(((((--((((....))-))).)))).)) ( -28.90) >DroEre_CAF1 4091 117 - 1 UUGGCAAUUUCCAACAUGCAACAUGUUGCACUGGAGUCAAGCCGA-AACCC-AAAACUUUUCCCGGGACUCCCGAAAUAAGAAAAAA-GUUGGGGAAAAAAAAUGUUUUUUUCGCCUGGC (((((...(((((...(((((....))))).)))))....)))))-.....-....(((....((((...))))....)))......-((..(((((((((.....))))))).))..)) ( -31.90) >DroYak_CAF1 3106 119 - 1 UUGGCAAUUUCCAACAUGCAACAUGUUGCACUGGAGUCAAACCCAAAACCCAAAAACUUUUCCCGGGACUCCCAAAAUAAGAAAAAAAAAUUGGAAAAAAAAAUGUUU-UUUCGCCUCGC ..(((..(((((((((((...)))))))....(((((....(((....................))))))))........)))).........(((((((.....)))-))))))).... ( -22.85) >consensus UUGGCAAUUUCCAACAUGCAACAUGUUGCACUGGAGUCAAGCCGA_AACCC_AAAACUUUUCCCGGGACUCCCGAAAAAGAAAAAAA_GUUGGGCA__AAAAAUGUUU_UUUCGCCUCGC ..(((....((((((.(((((....)))))..((((((...(((...................)))))))))................))))))..........(.......)))).... (-21.03 = -21.83 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:40 2006