
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,979,735 – 8,979,966 |
| Length | 231 |
| Max. P | 0.940342 |

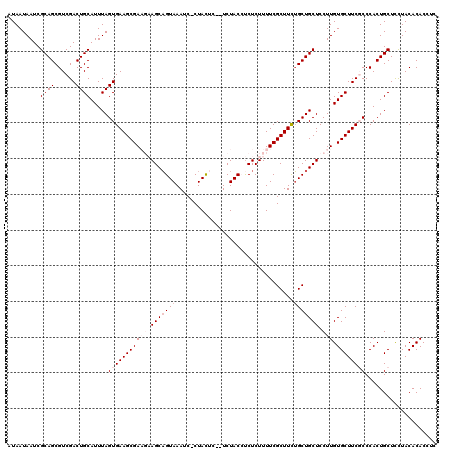
| Location | 8,979,735 – 8,979,852 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.41 |
| Mean single sequence MFE | -24.64 |
| Consensus MFE | -19.93 |
| Energy contribution | -21.53 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.940342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8979735 117 + 22224390 AUAAUAAUCGCAGCGUCGACUGCAUUUAGUGAAGCGAAGAAGCUGUAAAUC-CUACUC--UCUACCUCUCUUUUCGCUUCGGCUGCUCCUUGUGCUUCUCCCACUGCUCCUACACACCUC .........((((.(..((..((((.((((((((((((((((..(((....-......--..)))..)).)))))))))).))))......))))...)).).))))............. ( -24.10) >DroSec_CAF1 8293 119 + 1 AUAAUAAUCGCAGCGUCGACUGCAUUUAGUGAAGCGAAGAAGCAGUAAAUC-CUACUCUCUCUACCUCUCUUUUCGCUUCUGCUGCUCCUUGUGCUUCGCCCACUGCUCCUACACACCUC .........((((.(.(((..((((.((((((((((((((((..(((....-..........)))..)).)))))))))).))))......)))).)))..).))))............. ( -26.34) >DroSim_CAF1 3999 119 + 1 AUAAUAAUCGCAGCGUCGACUGCAUUUAGUGAAGCGAAGAAGCAGUAAAUC-CUACUCUCUCUACCUCUCUUUUCGCUUCUGCUGCUCCUUGUGCUUCGCCCACUGCUCCUACACACCUC .........((((.(.(((..((((.((((((((((((((((..(((....-..........)))..)).)))))))))).))))......)))).)))..).))))............. ( -26.34) >DroEre_CAF1 1787 114 + 1 AUAAUAAUCGCAGCGUCGACUGCAUUUAGUGAAGCGA---AGCAGUAAAUCCCUAUCC--UCUAC-UCUCUUUUCGCUUCUGCUGCUCCUUGUGCUUCGCCCACUGCUCCUACACACCUC .........((((.(.(((..((((.(((((((((((---((.((((...........--..)))-)....))))))))).))))......)))).)))..).))))............. ( -24.42) >DroYak_CAF1 569 113 + 1 AUAAUAAUCGCAGCGUCGACUGCAUUUAGUGAAGCGA---AGCAGUAAAUC-CUACUC--UCUAC-CCUCUUUUCGCUUUUGCUGCUCCUUGUGCUUCUCCCACUGCUUUUACACACCUC .........((((.(...((((....))))((((((.---(((((((((..-......--.....-............))))))))).....))))))...).))))............. ( -22.01) >consensus AUAAUAAUCGCAGCGUCGACUGCAUUUAGUGAAGCGAAGAAGCAGUAAAUC_CUACUC__UCUACCUCUCUUUUCGCUUCUGCUGCUCCUUGUGCUUCGCCCACUGCUCCUACACACCUC .........((((......)))).....(((((((((((.(((((((.................................))))))).))).)))))))).................... (-19.93 = -21.53 + 1.60)


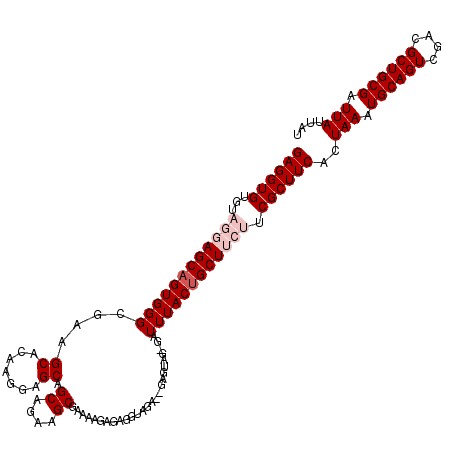
| Location | 8,979,735 – 8,979,852 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.41 |
| Mean single sequence MFE | -33.26 |
| Consensus MFE | -27.74 |
| Energy contribution | -29.14 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8979735 117 - 22224390 GAGGUGUGUAGGAGCAGUGGGAGAAGCACAAGGAGCAGCCGAAGCGAAAAGAGAGGUAGA--GAGUAG-GAUUUACAGCUUCUUCGCUUCACUAAAUGCAGUCGACGCUGCGAUUAUUAU .......((((..((((((...((.(((...((.....))((((((((..(((..(((((--......-..)))))..))).))))))))......)))..))..))))))..))))... ( -34.50) >DroSec_CAF1 8293 119 - 1 GAGGUGUGUAGGAGCAGUGGGCGAAGCACAAGGAGCAGCAGAAGCGAAAAGAGAGGUAGAGAGAGUAG-GAUUUACUGCUUCUUCGCUUCACUAAAUGCAGUCGACGCUGCGAUUAUUAU .......((((..((((((..(((.((.......)).((((((((((....(((((((((((......-..))).)))))))))))))))......)))..))).))))))..))))... ( -36.30) >DroSim_CAF1 3999 119 - 1 GAGGUGUGUAGGAGCAGUGGGCGAAGCACAAGGAGCAGCAGAAGCGAAAAGAGAGGUAGAGAGAGUAG-GAUUUACUGCUUCUUCGCUUCACUAAAUGCAGUCGACGCUGCGAUUAUUAU .......((((..((((((..(((.((.......)).((((((((((....(((((((((((......-..))).)))))))))))))))......)))..))).))))))..))))... ( -36.30) >DroEre_CAF1 1787 114 - 1 GAGGUGUGUAGGAGCAGUGGGCGAAGCACAAGGAGCAGCAGAAGCGAAAAGAGA-GUAGA--GGAUAGGGAUUUACUGCU---UCGCUUCACUAAAUGCAGUCGACGCUGCGAUUAUUAU .......((((..((((((..(((.((.......)).((((((((((......(-((((.--.(((....)))..)))))---)))))))......)))..))).))))))..))))... ( -31.50) >DroYak_CAF1 569 113 - 1 GAGGUGUGUAAAAGCAGUGGGAGAAGCACAAGGAGCAGCAAAAGCGAAAAGAGG-GUAGA--GAGUAG-GAUUUACUGCU---UCGCUUCACUAAAUGCAGUCGACGCUGCGAUUAUUAU .......((((..((((((...((.(((...(((((.((....)).........-...((--.(((((-......)))))---)))))))......)))..))..))))))..))))... ( -27.70) >consensus GAGGUGUGUAGGAGCAGUGGGCGAAGCACAAGGAGCAGCAGAAGCGAAAAGAGAGGUAGA__GAGUAG_GAUUUACUGCUUCUUCGCUUCACUAAAUGCAGUCGACGCUGCGAUUAUUAU ((((((...((((((((((((....((.......)).((....))..........................)))))))))))).))))))..(((.((((((....)))))).))).... (-27.74 = -29.14 + 1.40)

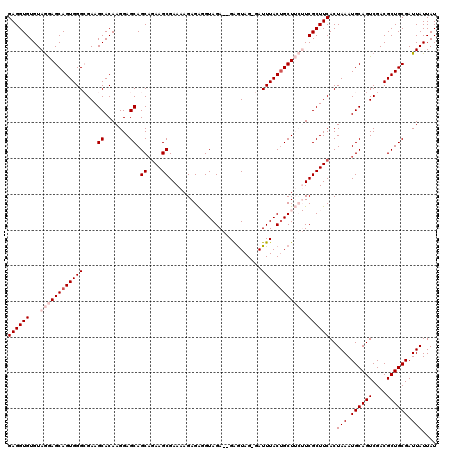
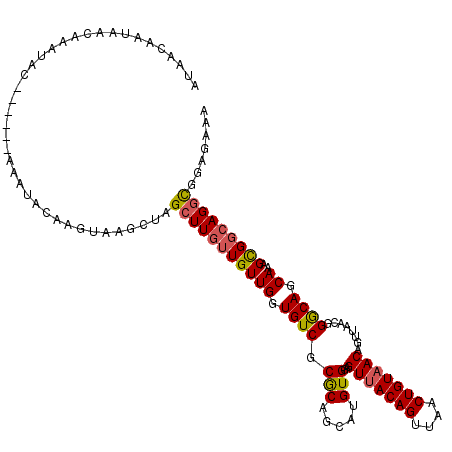
| Location | 8,979,852 – 8,979,966 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.45 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -24.65 |
| Energy contribution | -24.90 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8979852 114 - 22224390 AUAACAAUAACAAAUAC------AAAUACAAGUAAGCUAGCUUGUUGUUGGUGUCGCGCAGCAUGUGGAGUUACAGUUAACUGUAACAGUUAACGGACAGCAAAGCGGCAGGCGGAGAAA .................------.............((.(((((((((((.((((.(((.....)))..(((((((....)))))))........)))).))..)))))))))..))... ( -31.00) >DroSec_CAF1 8412 114 - 1 AUAACAAUAACAAAUAC------AAAUACAAGUAAGCUAGCUUGUUGUUGGUGUCGCACAGCAUGUGGAGUUACAGUUAACUGUAACAGUUAACGGGCAGCAAAGCGGCAGGCGGAGAAA .................------.............((.(((((((((((.((((.(((.....)))..(((((((....)))))))........)))).))..)))))))))..))... ( -30.80) >DroSim_CAF1 4118 120 - 1 AUAACAAUAACAAAUACAAAUACAAAUACAAGUAAGCUAGCUUGUUGUUGGUGUCGCGCAGCAUGUGGAGUUACAGUUAACUGUAACAGUUAACGGGCAGCAAAGCGGCAGGCGGAGAAA ....................................((.(((((((((((.((((.(((.....)))..(((((((....)))))))........)))).))..)))))))))..))... ( -30.40) >DroYak_CAF1 682 112 - 1 AUAACAAAAACAAAUAC------AAAUACAAAUAACAUAGGUUGUUGUUGUUGUCGCGCAGCAUGUGGAGUUACAGUUAACUGUUACAGUUAACGGGCAGCAACGUG--AGGUGGAGAAA ..............(((------...(((.((((((....))))))(((((((((.(((.....)))........((((((((...)))))))).))))))))))))--..)))...... ( -27.90) >consensus AUAACAAUAACAAAUAC______AAAUACAAGUAAGCUAGCUUGUUGUUGGUGUCGCGCAGCAUGUGGAGUUACAGUUAACUGUAACAGUUAACGGGCAGCAAAGCGGCAGGCGGAGAAA .......................................(((((((((((.((((.(((.....)))..(((((((....)))))))........)))).))..)))))))))....... (-24.65 = -24.90 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:33 2006