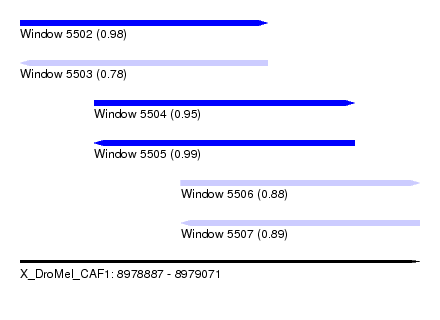
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,978,887 – 8,979,071 |
| Length | 184 |
| Max. P | 0.988048 |

| Location | 8,978,887 – 8,979,001 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.03 |
| Mean single sequence MFE | -28.91 |
| Consensus MFE | -23.58 |
| Energy contribution | -24.07 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
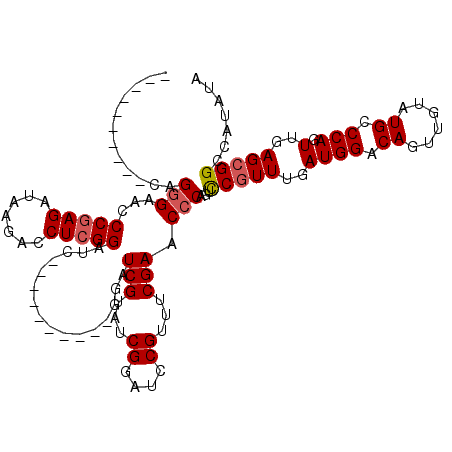
>X_DroMel_CAF1 8978887 114 + 22224390 UCUACAGCGAUCCUCCGACUUU------AUAUCAGCCAUAUAUUAUUUUUCGCUGUGUACACAGCGAAUGCCCUAUCGGUUAUAUGGCCCCUCAACUGGGCAUACAACUGUCCAUCAAAC ........(((...........------......((((((((......((((((((....)))))))).(((.....))))))))))).........(((((......)))))))).... ( -29.40) >DroSec_CAF1 7453 114 + 1 UCUACAUGGAUCCUCCGACUUU------AUGUCAGUCAUAUACUAUUUUUCGCUGUGUACACAGCGAAUGCCCUAUCGGUUAUAUGGCCGCUCAACUGGGCAUACAACUGUCCAUCAAAC .....((((((.....(((...------..))).((((((((......((((((((....)))))))).(((.....))))))))))).((((....))))........))))))..... ( -33.00) >DroSim_CAF1 2110 114 + 1 UCUACAUCGAUCCUCCGACUUU------AUGUCAGUCAUAUAUUAUUUUUCGCUGUGUACACAGCGAAUGCCCUAUCGGUUAUAUGGCCGCUCAACUGGGCAUACAACUGUCCAUCAAAC ................(((...------..))).((((((((......((((((((....)))))))).(((.....)))))))))))........((((((......))))))...... ( -29.50) >DroEre_CAF1 889 119 + 1 UCUACAUCGAUCCCCAAGCUGUAUAUGUAUAUCAGCCACAUAAU-UUUUUCGCUGUGUACAAAGCGAAUUCCCUAUCGGUUAUAUUGCCGCUCAACUGGGCAUACAACUGUCCAUCAAAC ........(((.....((.(((((.((((((.((((........-......))))))))))..((((((..((....))..)).)))).((((....))))))))).))....))).... ( -23.74) >consensus UCUACAUCGAUCCUCCGACUUU______AUAUCAGCCAUAUAUUAUUUUUCGCUGUGUACACAGCGAAUGCCCUAUCGGUUAUAUGGCCGCUCAACUGGGCAUACAACUGUCCAUCAAAC ........(((.......................((((((((......((((((((....))))))))...((....)).)))))))).........(((((......)))))))).... (-23.58 = -24.07 + 0.50)

| Location | 8,978,887 – 8,979,001 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.03 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -27.28 |
| Energy contribution | -27.34 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8978887 114 - 22224390 GUUUGAUGGACAGUUGUAUGCCCAGUUGAGGGGCCAUAUAACCGAUAGGGCAUUCGCUGUGUACACAGCGAAAAAUAAUAUAUGGCUGAUAU------AAAGUCGGAGGAUCGCUGUAGA .........(((((.((...((.........(((((((((.((....))...((((((((....))))))))......)))))))))(((..------...)))))...)).)))))... ( -34.60) >DroSec_CAF1 7453 114 - 1 GUUUGAUGGACAGUUGUAUGCCCAGUUGAGCGGCCAUAUAACCGAUAGGGCAUUCGCUGUGUACACAGCGAAAAAUAGUAUAUGACUGACAU------AAAGUCGGAGGAUCCAUGUAGA .....(((((.(((..(((((((.......(((........)))...)))).((((((((....)))))))).......)))..)))(((..------...)))......)))))..... ( -34.10) >DroSim_CAF1 2110 114 - 1 GUUUGAUGGACAGUUGUAUGCCCAGUUGAGCGGCCAUAUAACCGAUAGGGCAUUCGCUGUGUACACAGCGAAAAAUAAUAUAUGACUGACAU------AAAGUCGGAGGAUCGAUGUAGA .....(((..((((..(((((((.......(((........)))...)))).((((((((....)))))))).......)))..)))).)))------...((((......))))..... ( -33.60) >DroEre_CAF1 889 119 - 1 GUUUGAUGGACAGUUGUAUGCCCAGUUGAGCGGCAAUAUAACCGAUAGGGAAUUCGCUUUGUACACAGCGAAAAA-AUUAUGUGGCUGAUAUACAUAUACAGCUUGGGGAUCGAUGUAGA .........(((.(((.((.((((((((..((((.((((((((....))...((((((........))))))...-.)))))).))))(((....))).)))).)))).))))))))... ( -29.00) >consensus GUUUGAUGGACAGUUGUAUGCCCAGUUGAGCGGCCAUAUAACCGAUAGGGCAUUCGCUGUGUACACAGCGAAAAAUAAUAUAUGACUGACAU______AAAGUCGGAGGAUCGAUGUAGA ..........((((..(((((((.((((..............)))).)))).((((((((....)))))))).......)))..))))................................ (-27.28 = -27.34 + 0.06)



| Location | 8,978,921 – 8,979,041 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.52 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -28.95 |
| Energy contribution | -29.32 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8978921 120 + 22224390 UAUUAUUUUUCGCUGUGUACACAGCGAAUGCCCUAUCGGUUAUAUGGCCCCUCAACUGGGCAUACAACUGUCCAUCAAACGAAUUGGGUUCGAAACGGAUCCGAUACGAUCCGAUCCGAU ........((((((((....))))))))......(((((.....((..(((.....((((((......))))))((....))...)))..))...((((((......))))))..))))) ( -37.30) >DroSec_CAF1 7487 112 + 1 UACUAUUUUUCGCUGUGUACACAGCGAAUGCCCUAUCGGUUAUAUGGCCGCUCAACUGGGCAUACAACUGUCCAUCAAACGGACUGGGUUCGAAACGGAUCCGAUACGAUCC-------- ........((((((((....)))))))).((((....((((.((((.(((......))).)))).))))((((.......)))).)))).......(((((......)))))-------- ( -36.70) >DroSim_CAF1 2144 112 + 1 UAUUAUUUUUCGCUGUGUACACAGCGAAUGCCCUAUCGGUUAUAUGGCCGCUCAACUGGGCAUACAACUGUCCAUCAAACGGACUGGGUUCGAAACGGAUCCGAUACGAUCC-------- ........((((((((....)))))))).((((....((((.((((.(((......))).)))).))))((((.......)))).)))).......(((((......)))))-------- ( -36.70) >DroEre_CAF1 929 109 + 1 UAAU-UUUUUCGCUGUGUACAAAGCGAAUUCCCUAUCGGUUAUAUUGCCGCUCAACUGGGCAUACAACUGUCCAUCAAACGAACUGGGAUCGAAACGUUUCCGAUACGAU---------- ....-...((((((........)))))).((((..(((((......))).......((((((......))))))......))...))))......(((.......)))..---------- ( -25.00) >consensus UAUUAUUUUUCGCUGUGUACACAGCGAAUGCCCUAUCGGUUAUAUGGCCGCUCAACUGGGCAUACAACUGUCCAUCAAACGAACUGGGUUCGAAACGGAUCCGAUACGAUCC________ ........((((((((....)))))))).((((..(((((......))).......((((((......))))))......))...)))).......(((((......)))))........ (-28.95 = -29.32 + 0.37)


| Location | 8,978,921 – 8,979,041 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.52 |
| Mean single sequence MFE | -39.38 |
| Consensus MFE | -33.62 |
| Energy contribution | -33.88 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.988048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8978921 120 - 22224390 AUCGGAUCGGAUCGUAUCGGAUCCGUUUCGAACCCAAUUCGUUUGAUGGACAGUUGUAUGCCCAGUUGAGGGGCCAUAUAACCGAUAGGGCAUUCGCUGUGUACACAGCGAAAAAUAAUA .(((((.((((((......)))))).))))).(((...(((((.....))).(((((((((((.......))).)))))))).))..)))..((((((((....))))))))........ ( -44.90) >DroSec_CAF1 7487 112 - 1 --------GGAUCGUAUCGGAUCCGUUUCGAACCCAGUCCGUUUGAUGGACAGUUGUAUGCCCAGUUGAGCGGCCAUAUAACCGAUAGGGCAUUCGCUGUGUACACAGCGAAAAAUAGUA --------(((((......)))))........(((.((((((...)))))).((((((((.((........)).)))))))).....)))..((((((((....))))))))........ ( -39.80) >DroSim_CAF1 2144 112 - 1 --------GGAUCGUAUCGGAUCCGUUUCGAACCCAGUCCGUUUGAUGGACAGUUGUAUGCCCAGUUGAGCGGCCAUAUAACCGAUAGGGCAUUCGCUGUGUACACAGCGAAAAAUAAUA --------(((((......)))))........(((.((((((...)))))).((((((((.((........)).)))))))).....)))..((((((((....))))))))........ ( -39.80) >DroEre_CAF1 929 109 - 1 ----------AUCGUAUCGGAAACGUUUCGAUCCCAGUUCGUUUGAUGGACAGUUGUAUGCCCAGUUGAGCGGCAAUAUAACCGAUAGGGAAUUCGCUUUGUACACAGCGAAAAA-AUUA ----------.(((...((....))...)))((((.((((((...)))))).(((((((((((......).))).))))))).....)))).((((((........))))))...-.... ( -33.00) >consensus ________GGAUCGUAUCGGAUCCGUUUCGAACCCAGUCCGUUUGAUGGACAGUUGUAUGCCCAGUUGAGCGGCCAUAUAACCGAUAGGGCAUUCGCUGUGUACACAGCGAAAAAUAAUA ...........(((...((....))...))).(((.((((((...)))))).(((((((((((......).))).))))))).....)))..((((((((....))))))))........ (-33.62 = -33.88 + 0.25)

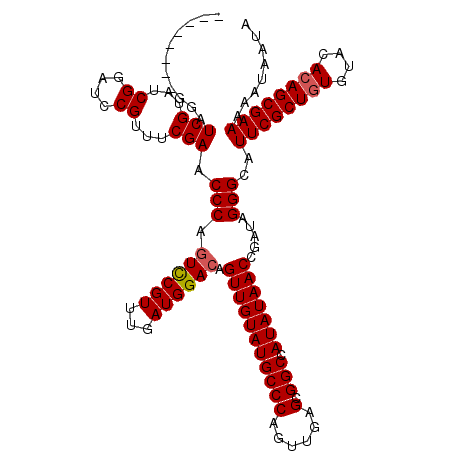
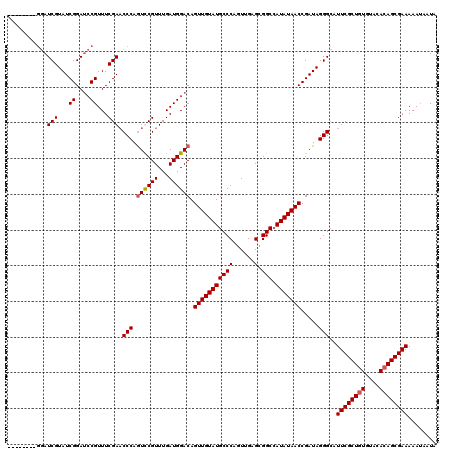
| Location | 8,978,961 – 8,979,071 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.18 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -22.36 |
| Energy contribution | -22.30 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8978961 110 + 22224390 UAUAUGGCCCCUCAACUGGGCAUACAACUGUCCAUCAAACGAAUUGGGUUCGAAACGGAUCCGAUACGAUCCGAUCCGAUCCGAUCCGAGGUCUUAUCUCGGGUUCCCGG---------- ...((((..((((...((((((......))))))......(.((((((((.((..((((((......)))))).)).)))))))).)))))..))))..(((....))).---------- ( -35.40) >DroSec_CAF1 7527 100 + 1 UAUAUGGCCGCUCAACUGGGCAUACAACUGUCCAUCAAACGGACUGGGUUCGAAACGGAUCCGAUACGAUCC----------GAUCCGAGGUCUUAUCUCGGGUUCCCUG---------- ..((((.(((......))).)))).....((((.......)))).(((..(....((((((......)))))----------)..(((((((...))))))))..)))..---------- ( -33.50) >DroSim_CAF1 2184 100 + 1 UAUAUGGCCGCUCAACUGGGCAUACAACUGUCCAUCAAACGGACUGGGUUCGAAACGGAUCCGAUACGAUCC----------GAUCCGAGGUCUUAUCUCGGGUUCCCUG---------- ..((((.(((......))).)))).....((((.......)))).(((..(....((((((......)))))----------)..(((((((...))))))))..)))..---------- ( -33.50) >DroEre_CAF1 968 105 + 1 UAUAUUGCCGCUCAACUGGGCAUACAACUGUCCAUCAAACGAACUGGGAUCGAAACGUUUCCGAUACGAU---------------CCCAGUUCUUAUCUCGGGUUCUCAGUUCUGAGUUC .........((((...((((((......))))))......((((((((((((...((....))...))))---------------)))))))).......)))).((((....))))... ( -34.80) >consensus UAUAUGGCCGCUCAACUGGGCAUACAACUGUCCAUCAAACGAACUGGGUUCGAAACGGAUCCGAUACGAUCC__________GAUCCGAGGUCUUAUCUCGGGUUCCCUG__________ ................((((((......))))))......((((((((((((...))))))))......................(((((((...))))))))))).............. (-22.36 = -22.30 + -0.06)

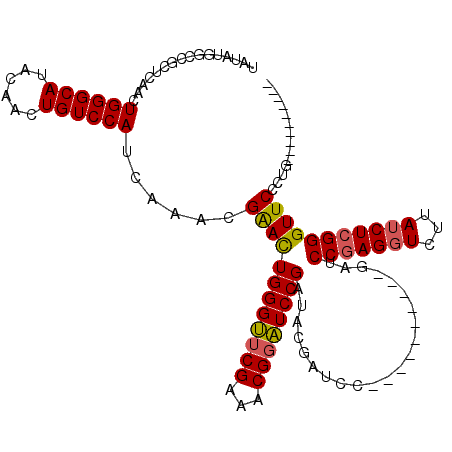
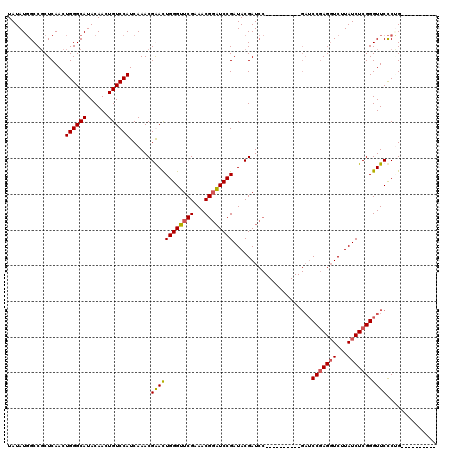
| Location | 8,978,961 – 8,979,071 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.18 |
| Mean single sequence MFE | -37.38 |
| Consensus MFE | -23.95 |
| Energy contribution | -24.45 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8978961 110 - 22224390 ----------CCGGGAACCCGAGAUAAGACCUCGGAUCGGAUCGGAUCGGAUCGUAUCGGAUCCGUUUCGAACCCAAUUCGUUUGAUGGACAGUUGUAUGCCCAGUUGAGGGGCCAUAUA ----------..(((...(((((.......))))).(((((.((((((.(((...))).)))))).))))).))).....(((.....)))...(((((((((.......))).)))))) ( -39.10) >DroSec_CAF1 7527 100 - 1 ----------CAGGGAACCCGAGAUAAGACCUCGGAUC----------GGAUCGUAUCGGAUCCGUUUCGAACCCAGUCCGUUUGAUGGACAGUUGUAUGCCCAGUUGAGCGGCCAUAUA ----------..(((...(((((.......)))))..(----------(((((......)))))).......))).(.(((((..((((.((......)).))).)..))))).)..... ( -34.50) >DroSim_CAF1 2184 100 - 1 ----------CAGGGAACCCGAGAUAAGACCUCGGAUC----------GGAUCGUAUCGGAUCCGUUUCGAACCCAGUCCGUUUGAUGGACAGUUGUAUGCCCAGUUGAGCGGCCAUAUA ----------..(((...(((((.......)))))..(----------(((((......)))))).......))).(.(((((..((((.((......)).))).)..))))).)..... ( -34.50) >DroEre_CAF1 968 105 - 1 GAACUCAGAACUGAGAACCCGAGAUAAGAACUGGG---------------AUCGUAUCGGAAACGUUUCGAUCCCAGUUCGUUUGAUGGACAGUUGUAUGCCCAGUUGAGCGGCAAUAUA ...((((....))))...((.......((((((((---------------((((...((....))...))))))))))))(((..((((.((......)).))).)..)))))....... ( -41.40) >consensus __________CAGGGAACCCGAGAUAAGACCUCGGAUC__________GGAUCGUAUCGGAUCCGUUUCGAACCCAGUCCGUUUGAUGGACAGUUGUAUGCCCAGUUGAGCGGCCAUAUA ............(((...(((((.......)))))................(((...((....))...))).)))...(((((..((((.((......)).))).)..)))))....... (-23.95 = -24.45 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:30 2006