
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,978,703 – 8,978,847 |
| Length | 144 |
| Max. P | 0.708600 |

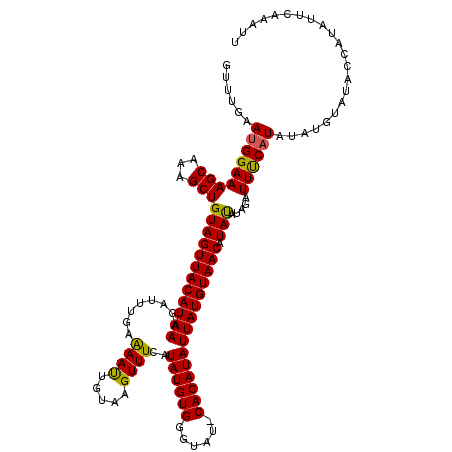
| Location | 8,978,703 – 8,978,810 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.19 |
| Mean single sequence MFE | -23.97 |
| Consensus MFE | -17.78 |
| Energy contribution | -17.23 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8978703 107 + 22224390 -----AAUGGAAAGCAAAGCUGUAGUUACAUAAAGAUUUGAAAACUGUAAGUUUCAUAUGUGGGUAUUCACAUAUUAUGUAACAUACAU--AUUCCAUG------UACCAUAUUCAAAUU -----.(((((((((...)))((((((((((((.....(((((........)))))(((((((....)))))))))))))))).)))..--.)))))).------............... ( -24.50) >DroSec_CAF1 7257 119 + 1 GUUUGAAAGGAAAGCAAAGCUGUAGUUACAUAAAGAUUGUAAAAUUGUAAGUUUCAUAUGUGGGUAU-CACAUAUUAUGUAACAUAUAUAGAUUUCAUAUAUGUAUACCAUAUUCAAAUU (((((((.((..............(((((((((.((((....))))..........((((((.....-)))))))))))))))(((((((.......)))))))...))...))))))). ( -22.30) >DroSim_CAF1 1914 119 + 1 GUUUGAAUGGAAAGCAAAGCUGUAGUUACAUAAAGAUUUGAGAAUUGUAAGUUUCAUAUGUGGGUAU-CACAUAUUAUGUAACAUAUAUAGAUUUCAUAUAUGUAUACCAUAUGCAAAUU (((((.((((..............(((((((((.....(((((........)))))((((((.....-)))))))))))))))(((((((.......)))))))...))))...))))). ( -25.10) >consensus GUUUGAAUGGAAAGCAAAGCUGUAGUUACAUAAAGAUUUGAAAAUUGUAAGUUUCAUAUGUGGGUAU_CACAUAUUAUGUAACAUAUAUAGAUUUCAUAUAUGUAUACCAUAUUCAAAUU ......(((((((((...)))((((((((((((........((((.....))))..((((((......))))))))))))))).))).....))))))...................... (-17.78 = -17.23 + -0.55)


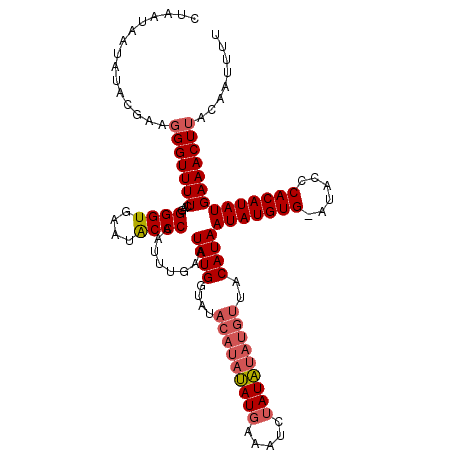
| Location | 8,978,738 – 8,978,847 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 86.86 |
| Mean single sequence MFE | -26.17 |
| Consensus MFE | -18.10 |
| Energy contribution | -19.77 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8978738 109 - 22224390 CGAAUAAUAUACGAAUGGUUUUCAGGGGUGAUAGCCCAAUUUGAAUAUGGUA------CAUGGAAU--AUGUAUGUUACAUAAUAUGUGAAUACCCACAUAUGAAACUUACAGUUUU ....(((((((((((.....)))..((((....))))...............------........--..))))))))....(((((((......)))))))(((((.....))))) ( -21.00) >DroSec_CAF1 7297 116 - 1 CUUAUAAUAUAGGAAGGGUUUUCAGGGGUGAAUACCCAAUUUGAAUAUGGUAUACAUAUAUGAAAUCUAUAUAUGUUACAUAAUAUGUG-AUACCCACAUAUGAAACUUACAAUUUU .........((((..((((.((((....)))).))))........((((....(((((((((.....)))))))))..))))(((((((-.....)))))))....))))....... ( -25.70) >DroSim_CAF1 1954 116 - 1 CUAAUAAUAUGCGAAGGGUUUUCAGGGGUGAAUACCCAAUUUGCAUAUGGUAUACAUAUAUGAAAUCUAUAUAUGUUACAUAAUAUGUG-AUACCCACAUAUGAAACUUACAAUUCU ......(((((((((((((.((((....)))).))))..))))))))).(((.(((((((((.....))))))))))))...(((((((-.....)))))))............... ( -31.80) >consensus CUAAUAAUAUACGAAGGGUUUUCAGGGGUGAAUACCCAAUUUGAAUAUGGUAUACAUAUAUGAAAUCUAUAUAUGUUACAUAAUAUGUG_AUACCCACAUAUGAAACUUACAAUUUU ...............(((((((...((((....))))........((((....(((((((((.....)))))))))..))))(((((((......))))))))))))))........ (-18.10 = -19.77 + 1.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:25 2006