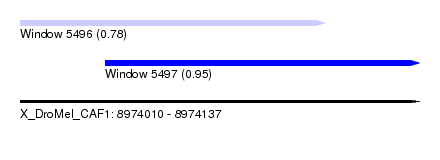
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,974,010 – 8,974,137 |
| Length | 127 |
| Max. P | 0.950384 |

| Location | 8,974,010 – 8,974,107 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.45 |
| Mean single sequence MFE | -19.69 |
| Consensus MFE | -12.85 |
| Energy contribution | -12.63 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8974010 97 + 22224390 ACUGCCA------CACCGAUGCUAUCCAGCA-------CAUCCAGCUCACUAAGCCCUUAAAUGCAUCCGUACUUUUGUGCAAAUGUAUAGCACGGCAACUACUUGCAGC .((((..------..(((.((((((.(((((-------......(((.....))).......)))....((((....))))...)).))))))))).........)))). ( -21.94) >DroEre_CAF1 19417 105 + 1 ACUGCCACCUCCACUCCGAUGCUAUCCAGCUAUCCAGCUAUCCAGCUCACUAAGCCCUUAAAUGCAUCCGUACU-UUGUGCAAAUGU--AGCACAGCAACUACUUGCA-- .................(((((.....((((....)))).....(((.....)))........)))))......-.(((((......--.)))))((((....)))).-- ( -19.40) >DroYak_CAF1 20113 85 + 1 A------------CUCCGAUGCUAUCCAGG--------CAUCCAGUUCACUAAGCCCUUAAAUGCAUCCGUACU-UUGUGCAAAUGU--AGCACAGCAACUACUUGCA-- .------------....(((((......((--------(....((....))..))).......)))))......-.(((((......--.)))))((((....)))).-- ( -17.72) >consensus ACUGCCA______CUCCGAUGCUAUCCAGC________CAUCCAGCUCACUAAGCCCUUAAAUGCAUCCGUACU_UUGUGCAAAUGU__AGCACAGCAACUACUUGCA__ .................(.((((.....................(((.....)))........((((..((((....))))..))))..))))).((((....))))... (-12.85 = -12.63 + -0.22)

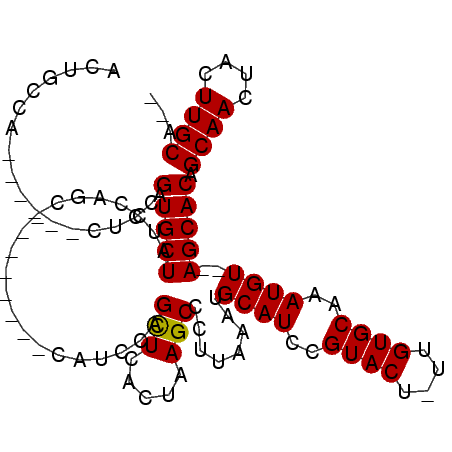
| Location | 8,974,037 – 8,974,137 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 93.90 |
| Mean single sequence MFE | -24.67 |
| Consensus MFE | -22.57 |
| Energy contribution | -22.13 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8974037 100 + 22224390 UCCAGCUCACUAAGCCCUUAAAUGCAUCCGUACUUUUGUGCAAAUGUAUAGCACGGCAACUACUUGCAGCUCUGGAUCGAUCAUCCAUCAUUCUGGGCAA .............((((....((((((..((((....))))..))))))(((...((((....)))).))).(((((.....))))).......)))).. ( -24.90) >DroEre_CAF1 19457 95 + 1 UCCAGCUCACUAAGCCCUUAAAUGCAUCCGUACU-UUGUGCAAAUGU--AGCACAGCAACUACUUGCA--UCUGGAUCGAUCAUCCAUCAUCCAGGGCAA .............((((....(((((...(((..-((((((......--.))))))....))).))))--).(((((.(((.....)))))))))))).. ( -25.30) >DroYak_CAF1 20133 95 + 1 UCCAGUUCACUAAGCCCUUAAAUGCAUCCGUACU-UUGUGCAAAUGU--AGCACAGCAACUACUUGCA--UCUGGAUCGAUCAUCCAUCAUCCUGGGCAA .............((((....(((((...(((..-((((((......--.))))))....))).))))--)..((((.(((.....))))))).)))).. ( -23.80) >consensus UCCAGCUCACUAAGCCCUUAAAUGCAUCCGUACU_UUGUGCAAAUGU__AGCACAGCAACUACUUGCA__UCUGGAUCGAUCAUCCAUCAUCCUGGGCAA .............((((.....((((...(((...((((((.........))))))....))).)))).....((((.(((.....))))))).)))).. (-22.57 = -22.13 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:20 2006