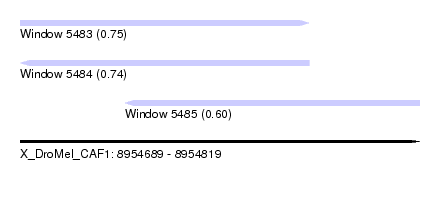
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,954,689 – 8,954,819 |
| Length | 130 |
| Max. P | 0.749013 |

| Location | 8,954,689 – 8,954,783 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 88.13 |
| Mean single sequence MFE | -31.88 |
| Consensus MFE | -22.00 |
| Energy contribution | -23.88 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8954689 94 + 22224390 -----CCACUGUG-CCAUCAAAUGCGCUAACGCCUAUCAGCGCGAGGCGUUAAUGCCUUUUGCGCACACUGCUUGUAAAUAUCGCGCUCUCGCUCGCACC -----.....(((-(..........((....)).....(((((((((((....)))).(((((((.....))..)))))..))))))).......)))). ( -25.90) >DroSec_CAF1 564 99 + 1 GACACCCACUGUG-CCACGAAACGCCCUAACGCCUAUCAGUUCGAGGCGUUAGUGCCUUUUGGGCACACUGCUUGUAAAUAUCGCGCUCUCGCUCGCACC .(((..((.((((-((.(((((.((.(((((((((.........))))))))).)).))))))))))).))..))).......(((........)))... ( -37.20) >DroSim_CAF1 600 99 + 1 GACACCCACUGUA-CCACCAAACGCACUAACGCCUAUCAGUUCGAGGCGUUAGUGCCUUUUGGGCACACUGCUUGUAAAUAUCGCGCUCUCGCUCGCACC .(((..((.(((.-...(((((.((((((((((((.........))))))))))))..)))))..))).))..))).......(((........)))... ( -31.90) >DroEre_CAF1 708 99 + 1 GAAAACCACUGUG-CGCCCAAAUGCGCUAACGCCUUUCAGCGCGACGCGUUAGUGCCUUUCGGGCACACUGCUUGUAAAUAUCGCGCUCUCGCUCGCACC ..........(((-((((((((.((((((((((...((.....)).)))))))))).))).))))..................(((....)))..)))). ( -33.60) >DroYak_CAF1 436 100 + 1 GACACCCACUGUGGCCCCCAAACGGGCUAACGCCUUCCUGCGCGAUGCGUUAGUGCUUUUUGGGCACACUGCUUGUAAAUAUCGCGCUCUCGCUCGCACC .........((((((........((((....))))....((((((((.....(((((.....)))))((.....))...))))))))....)).)))).. ( -30.80) >consensus GACACCCACUGUG_CCACCAAACGCGCUAACGCCUAUCAGCGCGAGGCGUUAGUGCCUUUUGGGCACACUGCUUGUAAAUAUCGCGCUCUCGCUCGCACC ......((.((((.(((..(((.((((((((((((.........)))))))))))).)))))).)))).))............(((........)))... (-22.00 = -23.88 + 1.88)

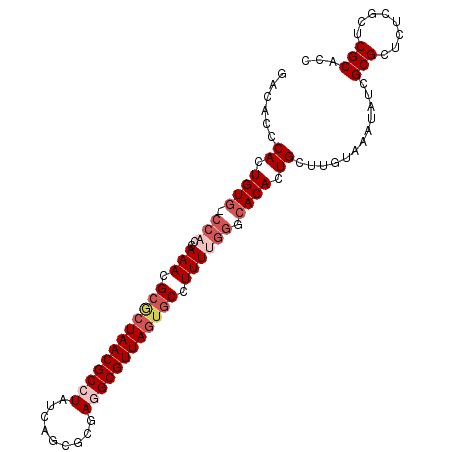

| Location | 8,954,689 – 8,954,783 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 88.13 |
| Mean single sequence MFE | -38.66 |
| Consensus MFE | -26.66 |
| Energy contribution | -27.34 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8954689 94 - 22224390 GGUGCGAGCGAGAGCGCGAUAUUUACAAGCAGUGUGCGCAAAAGGCAUUAACGCCUCGCGCUGAUAGGCGUUAGCGCAUUUGAUGG-CACAGUGG----- ....((.((....)).))...........((.(((((.((((((((......)))).((((((((....)))))))).))))...)-)))).)).----- ( -34.40) >DroSec_CAF1 564 99 - 1 GGUGCGAGCGAGAGCGCGAUAUUUACAAGCAGUGUGCCCAAAAGGCACUAACGCCUCGAACUGAUAGGCGUUAGGGCGUUUCGUGG-CACAGUGGGUGUC ....((.((....)).))......(((..((.(((((((.(((.((.(((((((((.........))))))))).)).))).).))-)))).))..))). ( -40.50) >DroSim_CAF1 600 99 - 1 GGUGCGAGCGAGAGCGCGAUAUUUACAAGCAGUGUGCCCAAAAGGCACUAACGCCUCGAACUGAUAGGCGUUAGUGCGUUUGGUGG-UACAGUGGGUGUC ....((.((....)).))......(((..((.(((((((.(((.((((((((((((.........)))))))))))).))).).))-)))).))..))). ( -43.20) >DroEre_CAF1 708 99 - 1 GGUGCGAGCGAGAGCGCGAUAUUUACAAGCAGUGUGCCCGAAAGGCACUAACGCGUCGCGCUGAAAGGCGUUAGCGCAUUUGGGCG-CACAGUGGUUUUC ....((.((....)).)).............(((((((((((.(((.(....).)))(((((((......))))))).))))))))-))).......... ( -40.10) >DroYak_CAF1 436 100 - 1 GGUGCGAGCGAGAGCGCGAUAUUUACAAGCAGUGUGCCCAAAAAGCACUAACGCAUCGCGCAGGAAGGCGUUAGCCCGUUUGGGGGCCACAGUGGGUGUC .((((........))))(((((((((.....(((.((((...((((.(((((((.((......))..)))))))...))))..))))))).))))))))) ( -35.10) >consensus GGUGCGAGCGAGAGCGCGAUAUUUACAAGCAGUGUGCCCAAAAGGCACUAACGCCUCGCGCUGAUAGGCGUUAGCGCGUUUGGUGG_CACAGUGGGUGUC ....((.((....)).))...........((.((((((..(((.((.(((((((((.........))))))))).)).)))...)).)))).))...... (-26.66 = -27.34 + 0.68)

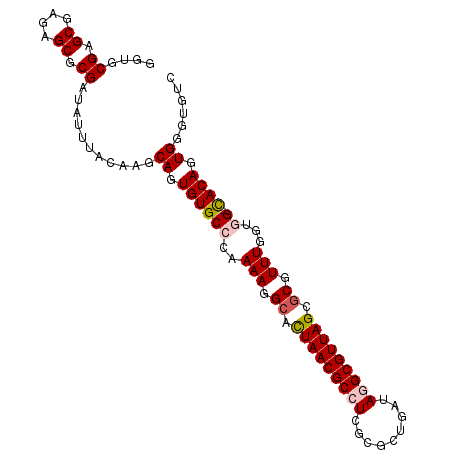
| Location | 8,954,723 – 8,954,819 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 96.35 |
| Mean single sequence MFE | -28.92 |
| Consensus MFE | -24.06 |
| Energy contribution | -24.70 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8954723 96 - 22224390 GAGAAUAUGUUGAUGAUGAGCGCAACAGUGGUAGUGGGUGCGAGCGAGAGCGCGAUAUUUACAAGCAGUGUGCGCAAAAGGCAUUAACGCCUCGCG (((....((((((((....(((((.((.((.(.(((((((((.((....)).)).))))))).).)).)))))))......)))))))).)))... ( -33.80) >DroSec_CAF1 603 96 - 1 GAGAAUAUGUUGAUGAUGAGCGCAACAGUGGUAGUGGGUGCGAGCGAGAGCGCGAUAUUUACAAGCAGUGUGCCCAAAAGGCACUAACGCCUCGAA (((....(((((.((.....)))))))(((...(((((((((.((....)).)).))))))).......(((((.....)))))...))))))... ( -28.10) >DroSim_CAF1 639 96 - 1 GAGAAUAUGUUGAUGAUGAGCGCAACAGUGGUAGUGGGUGCGAGCGAGAGCGCGAUAUUUACAAGCAGUGUGCCCAAAAGGCACUAACGCCUCGAA (((....(((((.((.....)))))))(((...(((((((((.((....)).)).))))))).......(((((.....)))))...))))))... ( -28.10) >DroEre_CAF1 747 96 - 1 GAGAAUAUGUUGAUGAUGAGCGCAACAGUGGUAGUGGGUGCGAGCGAGAGCGCGAUAUUUACAAGCAGUGUGCCCGAAAGGCACUAACGCGUCGCG .......(((((.((.....)))))))((((..(((((((((.((....)).)).)))))))..((...((((.(....)))))....)).)))). ( -28.50) >DroYak_CAF1 476 96 - 1 GAGAAUAUGUUGAUGAUGAGCGCAACAGUGGUAGUGGGUGCGAGCGAGAGCGCGAUAUUUACAAGCAGUGUGCCCAAAAAGCACUAACGCAUCGCG .......(((((.((.....)))))))(((((.(((((((((.((....)).)).)))))))..((...((((.......))))....))))))). ( -26.10) >consensus GAGAAUAUGUUGAUGAUGAGCGCAACAGUGGUAGUGGGUGCGAGCGAGAGCGCGAUAUUUACAAGCAGUGUGCCCAAAAGGCACUAACGCCUCGCG (((....(((((.((.....)))))))(((...(((((((((.((....)).)).))))))).......(((((.....)))))...))))))... (-24.06 = -24.70 + 0.64)

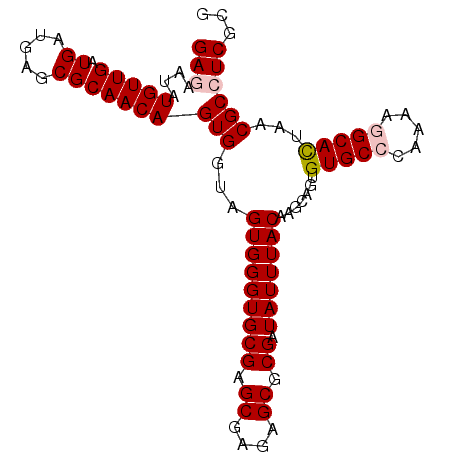

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:09 2006