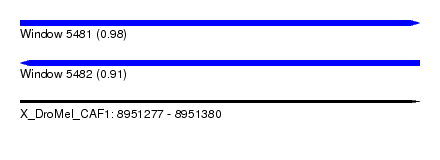
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,951,277 – 8,951,380 |
| Length | 103 |
| Max. P | 0.979013 |

| Location | 8,951,277 – 8,951,380 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 93.20 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -24.28 |
| Energy contribution | -24.17 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8951277 103 + 22224390 AACGUUUAACUUAAUGUGGUGGAAAUUGAAGCCUUCUCGCUGUACAAUCACCCACAUAUAAGAAGCUAAUGGUUAUUUCCCUUACGAGGACACCAUUACCAAC .........((((((((((.(...((((.(((......)))...))))..))))))).))))..(.(((((((....((((....).))).))))))).)... ( -22.20) >DroSec_CAF1 1550 103 + 1 AACGUUUAACAUAAUGUGGUGGAAAUUGAAGCCUUCUAGCUGUACCAUCAGCCACAUAUAUGAAGCUAAUGGUUAUUUCCUUUACGAGGAUACCAUUACCAAC .........(((((((((((.((......(((......)))......)).))))))).))))..(.(((((((....(((((...))))).))))))).)... ( -27.20) >DroSim_CAF1 1552 103 + 1 AAAUUAUAACAUAAUGUGGUGGAAAUUGAAGCCUUCUAGCUGUACGAUCAGCCACAUAUAUGAAGCUAAUGGUUAUUUCCUUUACGAGGACACCAUUACCAAC .........(((((((((((.((......(((......)))......)).))))))).))))..(.(((((((....(((((...))))).))))))).)... ( -25.80) >consensus AACGUUUAACAUAAUGUGGUGGAAAUUGAAGCCUUCUAGCUGUACAAUCAGCCACAUAUAUGAAGCUAAUGGUUAUUUCCUUUACGAGGACACCAUUACCAAC .........((((((((((..((......(((......)))......))..)))))).))))..(.(((((((....((((.....)))).))))))).)... (-24.28 = -24.17 + -0.11)



| Location | 8,951,277 – 8,951,380 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 93.20 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -25.86 |
| Energy contribution | -25.20 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8951277 103 - 22224390 GUUGGUAAUGGUGUCCUCGUAAGGGAAAUAACCAUUAGCUUCUUAUAUGUGGGUGAUUGUACAGCGAGAAGGCUUCAAUUUCCACCACAUUAAGUUAAACGUU ((((.(((((((.(((.(....))))....))))))).)..((((.((((((..(((((...(((......))).)))))....))))))))))...)))... ( -30.50) >DroSec_CAF1 1550 103 - 1 GUUGGUAAUGGUAUCCUCGUAAAGGAAAUAACCAUUAGCUUCAUAUAUGUGGCUGAUGGUACAGCUAGAAGGCUUCAAUUUCCACCACAUUAUGUUAAACGUU ..((((.((((.....))))...((((((.((((((((((.((....)).))))))))))..((((....))))...))))))))))................ ( -26.80) >DroSim_CAF1 1552 103 - 1 GUUGGUAAUGGUGUCCUCGUAAAGGAAAUAACCAUUAGCUUCAUAUAUGUGGCUGAUCGUACAGCUAGAAGGCUUCAAUUUCCACCACAUUAUGUUAUAAUUU ...(.(((((((.((((.....))))....))))))).)..((((.((((((..((......((((....))))......))..))))))))))......... ( -26.40) >consensus GUUGGUAAUGGUGUCCUCGUAAAGGAAAUAACCAUUAGCUUCAUAUAUGUGGCUGAUCGUACAGCUAGAAGGCUUCAAUUUCCACCACAUUAUGUUAAACGUU ...(.(((((((.((((.....))))....))))))).)..((((.((((((..((......(((......)))......))..))))))))))......... (-25.86 = -25.20 + -0.66)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:07 2006