
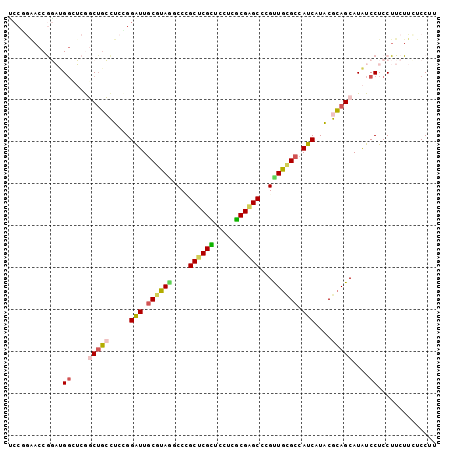
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,950,277 – 8,950,419 |
| Length | 142 |
| Max. P | 0.897523 |

| Location | 8,950,277 – 8,950,379 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.52 |
| Mean single sequence MFE | -33.95 |
| Consensus MFE | -23.28 |
| Energy contribution | -23.23 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8950277 102 + 22224390 UCCGGAACCAGAUGGCUCGGCUGCCUCCGGAUUGCGUAGUCCCGCUCGCUCUUCGCGAGCCCGCUGCGCCAUCAUACGCAGCAUAUCCUCCUUCUUCUCCUU ...(((...(((.((....(((((.....(((.(((((((...((((((.....))))))..))))))).)))....)))))....))....)))..))).. ( -39.50) >DroVir_CAF1 580 102 + 1 GCCAGAACCGCCGGGUGCAUCAGGCUCUGGAUUGCGUAUGCCCGCACGUUCCUCGCGUGCACGUUGCGCCAUCAUGCGCAGCAUAUCCUCCUUCUUCUCCUU .(((((...(((.((....)).))))))))...(.((((((..((((((.....))))))....(((((......))))))))))).).............. ( -32.10) >DroSec_CAF1 586 102 + 1 UCCGGAACCGGAUGUCUCGGCUGCCUCCGGAUUGCGUAGUCCCGCUCGCUCUUCGCGAGCCCGUUGCGCCAUCAUACGCAGCAUAUCCUCCUUCUUCUCCUU ...(((...((((((....(((((.....(((.((((((.(..((((((.....))))))..))))))).)))....))))))))))))))........... ( -36.60) >DroEre_CAF1 586 102 + 1 UCCUAGUCCGGAUGGUUCGGCUGCCUCCGGAUUUCGUAGUCCUGCUCGCUCUUCGCGAGCCCGCUGUGCCAUCAUACGCAGCAUAUCCUCCUUCUUCUCCUU ....((((((((.(((......)))))))))))..........((((((.....))))))..((((((........)))))).................... ( -34.40) >DroMoj_CAF1 580 96 + 1 ------ACCACCCGGUGCAUCUGGUUCGGGAUUGCGUAAGCCAGCACGUUCCUCGCGUGCACGUUGCGCCAUCAUACGCAGCAUGUCCUCCUUCUUCUCCUU ------.....((((.....))))...(((((.((....))..((((((.....))))))..((((((........))))))..)))))............. ( -27.80) >DroAna_CAF1 586 102 + 1 CCCCGAACCGGAUGGCUCGGCUCCCUCGGGGUUGCGCAAGCCAGCUCGUUCCUCACGUGCCCGUUGGGCCAUCAUGCGGAGCAUAUCCUCCUUCUUUUCCUU ....(((...((((((((((((((....)))..((....))..((.(((.....))).))..)))))))))))....((((......)))))))........ ( -33.30) >consensus UCCGGAACCGGAUGGCUCGGCUGCCUCCGGAUUGCGUAGGCCCGCUCGCUCCUCGCGAGCCCGUUGCGCCAUCAUACGCAGCAUAUCCUCCUUCUUCUCCUU .............((....(((((.....(((.((((((....((((((.....))))))...)))))).)))....)))))....)).............. (-23.28 = -23.23 + -0.05)


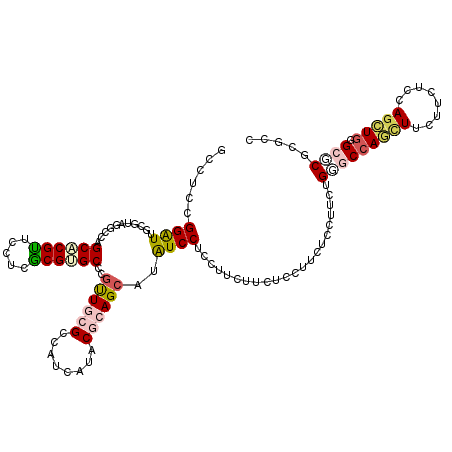
| Location | 8,950,299 – 8,950,419 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -36.01 |
| Consensus MFE | -22.10 |
| Energy contribution | -23.05 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8950299 120 + 22224390 GCCUCCGGAUUGCGUAGUCCCGCUCGCUCUUCGCGAGCCCGCUGCGCCAUCAUACGCAGCAUAUCCUCCUUCUUCUCCUUCUCCUUUUGGGCCAGCUUCUUUUCCAGCUGGGAGCGUGCC ((.....(((.(((((((...((((((.....))))))..))))))).)))....)).((((...((((............(((....))).(((((........))))))))).)))). ( -42.10) >DroVir_CAF1 602 120 + 1 GGCUCUGGAUUGCGUAUGCCCGCACGUUCCUCGCGUGCACGUUGCGCCAUCAUGCGCAGCAUAUCCUCCUUCUUCUCCUUCUCUUUCUGUGCCAGCUUCUUCUCCAACUGAGCACGCGCC (((.(.(((..(.((((((..((((((.....))))))....(((((......))))))))))).))))...................(((((((............))).))))).))) ( -35.60) >DroSec_CAF1 608 120 + 1 GCCUCCGGAUUGCGUAGUCCCGCUCGCUCUUCGCGAGCCCGUUGCGCCAUCAUACGCAGCAUAUCCUCCUUCUUCUCCUUCUCCUUUUGGGCAAGCUUCUUUUCCAGCUGGGAGCGUGCC ((.....(((.((((((.(..((((((.....))))))..))))))).)))....)).((((...(((((...........(((....)))..((((........))))))))).)))). ( -35.60) >DroWil_CAF1 614 120 + 1 ACUUCAGGAUUCCGUAGUCCAGCACGUUCUUCGCGGGCUCUUUGGGCCAUUAAACGAAGCAUAUCCUCUUUUUUCUCCUUCUCCUUUUGGGCCAAUUUUUUUUCAAGUUGGGCUCGCGCC ......((((......))))...........(((((((((.((((.(((......((((..................))))......))).))))..............))))))))).. ( -28.33) >DroMoj_CAF1 596 120 + 1 GGUUCGGGAUUGCGUAAGCCAGCACGUUCCUCGCGUGCACGUUGCGCCAUCAUACGCAGCAUGUCCUCCUUCUUCUCCUUCUCUUUCUGCGCCAACUUCUUCUCCAGCUGGGCGCGAGCU ((((((((((.((....))..((((((.....))))))..((((((........))))))..))))).....................(((((..((........))...)))))))))) ( -38.30) >DroAna_CAF1 608 120 + 1 CCCUCGGGGUUGCGCAAGCCAGCUCGUUCCUCACGUGCCCGUUGGGCCAUCAUGCGGAGCAUAUCCUCCUUCUUUUCCUUCUCCUUCUGAGCCAGUUUCUUCUCCAGCUGGGCGCGCGCU ......(((((((....).))))))........(((((((((((((.......((((((......))))...........(((.....)))...))......)))))).))))))).... ( -36.12) >consensus GCCUCCGGAUUGCGUAGGCCAGCACGUUCCUCGCGUGCCCGUUGCGCCAUCAUACGCAGCAUAUCCUCCUUCUUCUCCUUCUCCUUCUGGGCCAGCUUCUUCUCCAGCUGGGCGCGCGCC ......((((...........((((((.....))))))..((((((........))))))..))))......................(((((((((........))))).))))..... (-22.10 = -23.05 + 0.95)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:05 2006