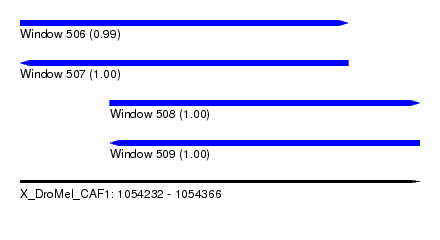
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,054,232 – 1,054,366 |
| Length | 134 |
| Max. P | 0.999964 |

| Location | 1,054,232 – 1,054,342 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.77 |
| Mean single sequence MFE | -33.74 |
| Consensus MFE | -31.07 |
| Energy contribution | -32.15 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1054232 110 + 22224390 ----------UGAAAAUGCGAAUCUGCCAGUGAGUGCGGAAGCUGCCGACCGCUUUUUAAUUUGAGUGGAAAGUAGUCGACAGGCGCUGCCGCAUUUCAAAAUUAUAGAAAUUGGCACAA ----------..............(((((((((((((((.(((.((((((..(((((((.......)))))))..)))....)))))).)))))))).............)))))))... ( -35.01) >DroSec_CAF1 3177 110 + 1 ----------UGGAAAUGCGAAUCUGCCAGUGAGUGCGGAAGCUGCCGACCGCUUUUUAAUUUGAGUGGAAAGUAGUCGACAGGCGCUGCCGCAUUUCAAAAUUAUAGAAAUUGGCACAA ----------..............(((((((((((((((.(((.((((((..(((((((.......)))))))..)))....)))))).)))))))).............)))))))... ( -35.01) >DroSim_CAF1 4112 110 + 1 ----------UGGAAAUGCGAAUCUGCCAGUGAGUGCGGAAGCUGCCGACCGCUUUUUAAUUUGAAUGGAAAGUAGUCGACAGGCGCUGCCGCAUUUCAAAAUUAUAGAAAUUGGCACAA ----------..............(((((((((((((((.(((.((((((..(((((((.......)))))))..)))....)))))).)))))))).............)))))))... ( -35.01) >DroEre_CAF1 3198 109 + 1 GG------AGUGGGAAUGGGAAUCUGCCA--GAGUGCGGAAGCUGCCGACCGCUUUUUAAUUUGAGUGGAAAG---UCGACAGGCGCUGCCGCAUUUCAAAAUUAUAGAAAUUGGCACAA ..------................(((((--((((((((.(((.(((..((((((........))))))...(---....).)))))).))))))))...............)))))... ( -31.76) >DroYak_CAF1 5339 117 + 1 GAAACUGCAAUGGGAAUGGGAAUCUGCCAAAGAGUGCGGAAGCUGCCAACCGCUUUUUAAUUUGAGUGGAAAG---UCGACAGGCGCUGCCGCAUUUCAAAAUUAUAGAAAUUGGCACAA ........................((((((...((((((.(((.(((..((((((........))))))...(---....).)))))).))))))(((.........))).))))))... ( -31.90) >consensus __________UGGAAAUGCGAAUCUGCCAGUGAGUGCGGAAGCUGCCGACCGCUUUUUAAUUUGAGUGGAAAGUAGUCGACAGGCGCUGCCGCAUUUCAAAAUUAUAGAAAUUGGCACAA ........................(((((((((((((((.(((.(((..((((((........))))))...((.....)).)))))).)))))))).............)))))))... (-31.07 = -32.15 + 1.08)

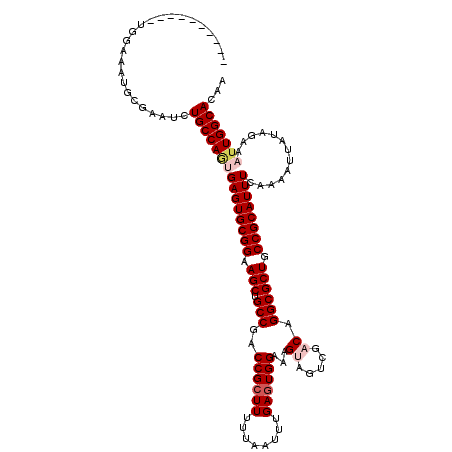
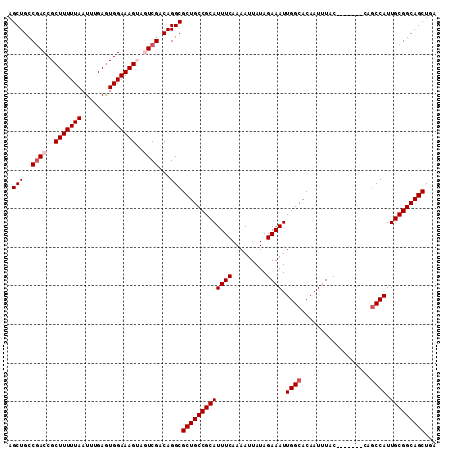
| Location | 1,054,232 – 1,054,342 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.77 |
| Mean single sequence MFE | -31.24 |
| Consensus MFE | -28.94 |
| Energy contribution | -29.58 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.35 |
| SVM RNA-class probability | 0.999878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1054232 110 - 22224390 UUGUGCCAAUUUCUAUAAUUUUGAAAUGCGGCAGCGCCUGUCGACUACUUUCCACUCAAAUUAAAAAGCGGUCGGCAGCUUCCGCACUCACUGGCAGAUUCGCAUUUUCA---------- .(.((((((((((.........)))))((((.(((...(((((((..((((.............))))..)))))))))).))))......))))).)............---------- ( -32.32) >DroSec_CAF1 3177 110 - 1 UUGUGCCAAUUUCUAUAAUUUUGAAAUGCGGCAGCGCCUGUCGACUACUUUCCACUCAAAUUAAAAAGCGGUCGGCAGCUUCCGCACUCACUGGCAGAUUCGCAUUUCCA---------- .(.((((((((((.........)))))((((.(((...(((((((..((((.............))))..)))))))))).))))......))))).)............---------- ( -32.32) >DroSim_CAF1 4112 110 - 1 UUGUGCCAAUUUCUAUAAUUUUGAAAUGCGGCAGCGCCUGUCGACUACUUUCCAUUCAAAUUAAAAAGCGGUCGGCAGCUUCCGCACUCACUGGCAGAUUCGCAUUUCCA---------- .(.((((((((((.........)))))((((.(((...(((((((..((((.............))))..)))))))))).))))......))))).)............---------- ( -32.32) >DroEre_CAF1 3198 109 - 1 UUGUGCCAAUUUCUAUAAUUUUGAAAUGCGGCAGCGCCUGUCGA---CUUUCCACUCAAAUUAAAAAGCGGUCGGCAGCUUCCGCACUC--UGGCAGAUUCCCAUUCCCACU------CC .(.((((((((((.........)))))((((.((((((..(((.---((((.............)))))))..))).))).))))....--))))).)..............------.. ( -29.42) >DroYak_CAF1 5339 117 - 1 UUGUGCCAAUUUCUAUAAUUUUGAAAUGCGGCAGCGCCUGUCGA---CUUUCCACUCAAAUUAAAAAGCGGUUGGCAGCUUCCGCACUCUUUGGCAGAUUCCCAUUCCCAUUGCAGUUUC .(.((((((((((.........))))(((((.((((((..(((.---((((.............)))))))..))).))).)))))....)))))).)...................... ( -29.82) >consensus UUGUGCCAAUUUCUAUAAUUUUGAAAUGCGGCAGCGCCUGUCGACUACUUUCCACUCAAAUUAAAAAGCGGUCGGCAGCUUCCGCACUCACUGGCAGAUUCGCAUUUCCA__________ .(.((((((((((.........)))))((((.(((...((((((((.((((.............)))).))))))))))).))))......))))).)...................... (-28.94 = -29.58 + 0.64)

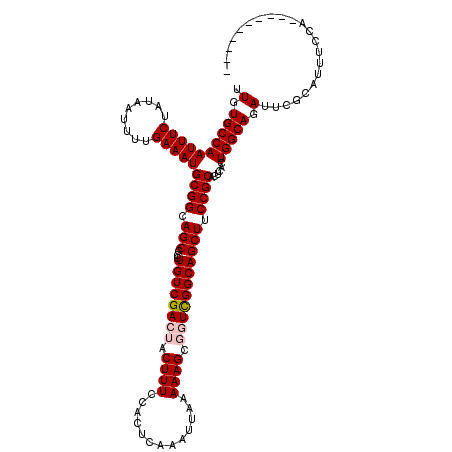
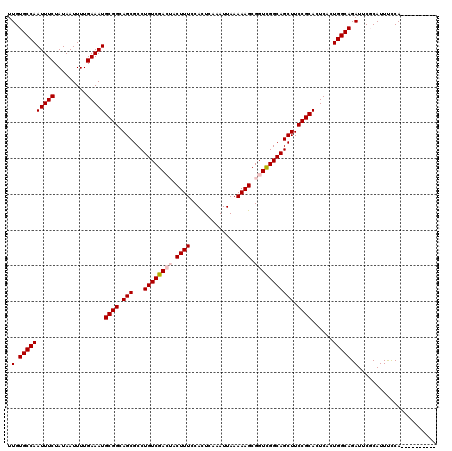
| Location | 1,054,262 – 1,054,366 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 94.18 |
| Mean single sequence MFE | -36.42 |
| Consensus MFE | -32.33 |
| Energy contribution | -33.13 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.57 |
| Structure conservation index | 0.89 |
| SVM decision value | 4.56 |
| SVM RNA-class probability | 0.999920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1054262 104 + 22224390 AGCUGCCGACCGCUUUUUAAUUUGAGUGGAAAGUAGUCGACAGGCGCUGCCGCAUUUCAAAAUUAUAGAAAUUGGCACAAUUUAC-------CAUCCAUUGCGGCAGCUGA .(((..((((..(((((((.......)))))))..))))...)))(((((((((((((.........)))).(((.........)-------)).....)))))))))... ( -35.00) >DroSec_CAF1 3207 104 + 1 AGCUGCCGACCGCUUUUUAAUUUGAGUGGAAAGUAGUCGACAGGCGCUGCCGCAUUUCAAAAUUAUAGAAAUUGGCACAAUUUAC-------CAGCCAUUGCGGCAGCUGA .(((..((((..(((((((.......)))))))..))))...)))(((((((((((((.........)))).((((.........-------..)))).)))))))))... ( -38.60) >DroSim_CAF1 4142 104 + 1 AGCUGCCGACCGCUUUUUAAUUUGAAUGGAAAGUAGUCGACAGGCGCUGCCGCAUUUCAAAAUUAUAGAAAUUGGCACAAUUUAC-------CAGCCAUUGCGGCAGCUGA .(((..((((..(((((((.......)))))))..))))...)))(((((((((((((.........)))).((((.........-------..)))).)))))))))... ( -38.60) >DroEre_CAF1 3230 108 + 1 AGCUGCCGACCGCUUUUUAAUUUGAGUGGAAAG---UCGACAGGCGCUGCCGCAUUUCAAAAUUAUAGAAAUUGGCACAAUUUACCAGUUACCAGCCAUUGCGGCAGCUGA ((((((((.((((((........))))))....---......((((.(((((.(((((.........)))))))))))(((......)))....)))....)))))))).. ( -35.50) >DroYak_CAF1 5379 101 + 1 AGCUGCCAACCGCUUUUUAAUUUGAGUGGAAAG---UCGACAGGCGCUGCCGCAUUUCAAAAUUAUAGAAAUUGGCACAAUUUAC-------CAGCCAUUGCGGCAGCUUA ....(((..((((((........))))))...(---....).)))(((((((((((((.........)))).((((.........-------..)))).)))))))))... ( -34.40) >consensus AGCUGCCGACCGCUUUUUAAUUUGAGUGGAAAGUAGUCGACAGGCGCUGCCGCAUUUCAAAAUUAUAGAAAUUGGCACAAUUUAC_______CAGCCAUUGCGGCAGCUGA .(((..((((..(((((((.......)))))))..))))...)))(((((((((((((.........)))).((((..................)))).)))))))))... (-32.33 = -33.13 + 0.80)

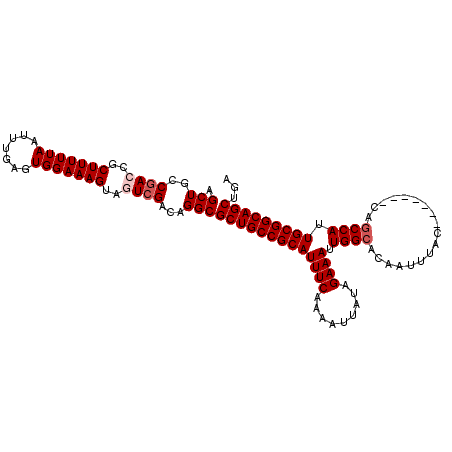
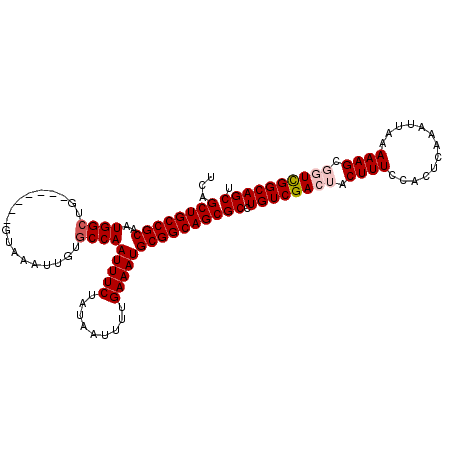
| Location | 1,054,262 – 1,054,366 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 94.18 |
| Mean single sequence MFE | -38.29 |
| Consensus MFE | -35.33 |
| Energy contribution | -36.17 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.18 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.96 |
| SVM RNA-class probability | 0.999964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1054262 104 - 22224390 UCAGCUGCCGCAAUGGAUG-------GUAAAUUGUGCCAAUUUCUAUAAUUUUGAAAUGCGGCAGCGCCUGUCGACUACUUUCCACUCAAAUUAAAAAGCGGUCGGCAGCU ...((((((((......((-------(((.....)))))(((((.........)))))))))))))((.(((((((..((((.............))))..))))))))). ( -39.92) >DroSec_CAF1 3207 104 - 1 UCAGCUGCCGCAAUGGCUG-------GUAAAUUGUGCCAAUUUCUAUAAUUUUGAAAUGCGGCAGCGCCUGUCGACUACUUUCCACUCAAAUUAAAAAGCGGUCGGCAGCU ...((((((((..((((..-------.........))))(((((.........)))))))))))))((.(((((((..((((.............))))..))))))))). ( -40.52) >DroSim_CAF1 4142 104 - 1 UCAGCUGCCGCAAUGGCUG-------GUAAAUUGUGCCAAUUUCUAUAAUUUUGAAAUGCGGCAGCGCCUGUCGACUACUUUCCAUUCAAAUUAAAAAGCGGUCGGCAGCU ...((((((((..((((..-------.........))))(((((.........)))))))))))))((.(((((((..((((.............))))..))))))))). ( -40.52) >DroEre_CAF1 3230 108 - 1 UCAGCUGCCGCAAUGGCUGGUAACUGGUAAAUUGUGCCAAUUUCUAUAAUUUUGAAAUGCGGCAGCGCCUGUCGA---CUUUCCACUCAAAUUAAAAAGCGGUCGGCAGCU ...((((((((..((((..................))))(((((.........)))))))))))))((.((((((---((....................)))))))))). ( -36.02) >DroYak_CAF1 5379 101 - 1 UAAGCUGCCGCAAUGGCUG-------GUAAAUUGUGCCAAUUUCUAUAAUUUUGAAAUGCGGCAGCGCCUGUCGA---CUUUCCACUCAAAUUAAAAAGCGGUUGGCAGCU ...((((((((..((((..-------.........))))(((((.........)))))))))))))((.((((((---((....................)))))))))). ( -34.45) >consensus UCAGCUGCCGCAAUGGCUG_______GUAAAUUGUGCCAAUUUCUAUAAUUUUGAAAUGCGGCAGCGCCUGUCGACUACUUUCCACUCAAAUUAAAAAGCGGUCGGCAGCU ...((((((((..((((..................))))(((((.........)))))))))))))((.((((((((.((((.............)))).)))))))))). (-35.33 = -36.17 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:01 2006