
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,922,073 – 8,922,176 |
| Length | 103 |
| Max. P | 0.862287 |

| Location | 8,922,073 – 8,922,176 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.33 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -12.59 |
| Energy contribution | -13.73 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
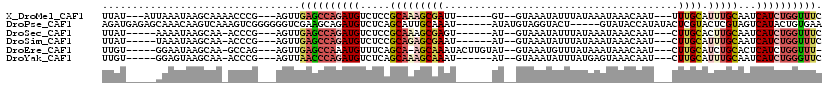
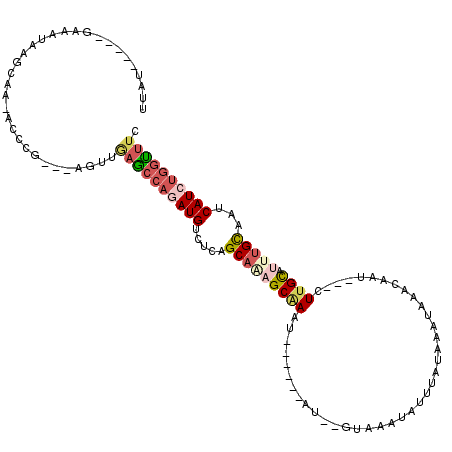
>X_DroMel_CAF1 8922073 103 + 22224390 UUAU---AUUAAAUAAGCAAAACCCG---AGUUGAGCCAGAUGUCUCCGCAAAGCGAUU------GU--GUAAAUAUUUAUAAAUAAACAAU---UUUGCAUUUGCAAUCAUCUGGUUUC ....---...................---....((((((((((.....(((((((((..------.(--((...((((....)))).)))..---.)))).)))))...)))))))))). ( -23.00) >DroPse_CAF1 53091 109 + 1 AGAUGAGAGCAAACAAGUCAAAGUCGGGGGGUCGAAGCAGAUGUCUCAGCAUUGCAAAU------AUAUGUAGGUACU-----GUAUACCAUAUACUCGUACUCGUAGUCAUACUGUGAA ..(((((.........(.(((.((.((((.(((......))).)))).)).))))....------.(((((.((((..-----...))))))))))))))..((((((.....)))))). ( -22.80) >DroSec_CAF1 36059 100 + 1 UUAU-----AAAAUAAGCAA-ACCCG---AGUUGAGCCAGAUGUCUCCGCAAAGCGAGU------AU--GUAAAUAUUUAUAAAUAAACAAU---CUUGCACUUGCAAUCAUCUGGUUUC ....-----...........-.....---....((((((((((.....((((.(((((.------.(--((...((((....)))).)))..---)))))..))))...)))))))))). ( -22.40) >DroSim_CAF1 32404 100 + 1 UUAU-----UAAAUAAGCAA-ACCCG---AGUUGAGCCAGAUGUCUCCGCAGAGCGAAU------AU--GUAAAUAUUUAUAAAUAAACAAU---CUUGCAUUUGCAAUCAUCUGGUUUC ....-----...........-.....---....((((((((((.....(((((((((..------.(--((...((((....)))).)))..---.)))).)))))...)))))))))). ( -21.30) >DroEre_CAF1 36216 104 + 1 UUGU-----GGAAUAAGCAA-GCCAG---AGUUGAGCCAAAUGUUUCAGCA-AGCAAAUACUUGUAU--GUAAAUGUUUAUAAAUAAACAAU---CUUGCAUCUGCACUCAUCUGGUUU- ....-----.........((-(((((---(((((((.(....).)))))).-.((((.(((......--)))..((((((....))))))..---.))))...........))))))))- ( -22.40) >DroYak_CAF1 37066 100 + 1 UUGU-----GGAGUAAGCAA-ACCCG---AGUUAACCCAGAUGUCUCAGCAAAGCAAAU------AU--GUAAAUAUUUAUGAGUAAACAAU---CUUGCAUUUGCAAUCAUCUGGGUUC ...(-----((.((......-)))))---....((((((((((.....(((((((((((------((--....))))))..(((........---))))).)))))...)))))))))). ( -26.10) >consensus UUAU_____GAAAUAAGCAA_ACCCG___AGUUGAGCCAGAUGUCUCAGCAAAGCAAAU______AU__GUAAAUAUUUAUAAAUAAACAAU___CUUGCAUUUGCAAUCAUCUGGUUUC .................................((((((((((.....(((((((((.......................................)))).)))))...)))))))))). (-12.59 = -13.73 + 1.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:51 2006