
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,921,661 – 8,921,870 |
| Length | 209 |
| Max. P | 0.999898 |

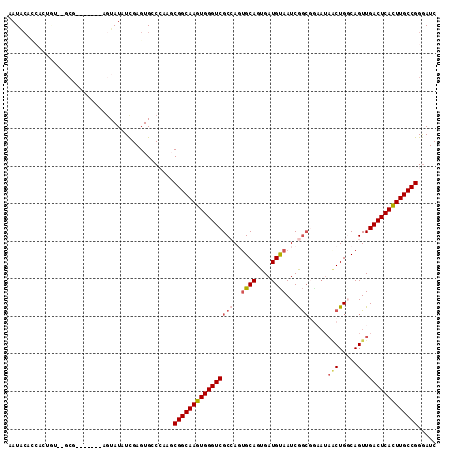
| Location | 8,921,661 – 8,921,768 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 74.93 |
| Mean single sequence MFE | -42.74 |
| Consensus MFE | -26.54 |
| Energy contribution | -28.96 |
| Covariance contribution | 2.42 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.77 |
| Structure conservation index | 0.62 |
| SVM decision value | 4.44 |
| SVM RNA-class probability | 0.999898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8921661 107 + 22224390 AAUGCAUCAGUGUACGCG-------AGUAUAUCGAGUGCCCAAGCGGCAAGUGGGUCGCCAGUGCAGUGAUGUAAUCGGCCAAAUAACUGCCAGUUGACUCACUUGCCGGAAUC ...(((((.((((((...-------.)))))).)).))).....((((((((((((((((..((((....))))...))).....(((.....))))))))))))))))..... ( -44.70) >DroSec_CAF1 35654 97 + 1 AAUACACCACUGU--GCG-------AGUAUAUCGAGUGGCCAAGCGGCAAGUGGGUCGCCAGUGCAGUGAUGUAAUCGG--------CUGGCAGUUGACUCACUUGCCGGGAUC ......(((((..--.((-------(.....)))))))).....((((((((((((((((((((((....))).....)--------)))))....)))))))))))))..... ( -41.80) >DroSim_CAF1 31993 105 + 1 AAUACACCACUUU--GCG-------AGUAUAUCGAGUGGCCAAGCGGCAAGUGGGUCGCCAGUGCAGUGAUGUAAUCGGCGGAAUAGCUGGCAGUUGACUCACUUGCCGGGAUC ......(((((..--.((-------(.....)))))))).....((((((((((((((((((((((....)))..((....))...))))))....)))))))))))))..... ( -42.10) >DroEre_CAF1 35856 93 + 1 AAU----------------ACACACCGUAUGUUGAGUGCC----CGGCAAGUGGGUCGC-AUGGCAGUGAUGCAAUCAACGGAAUAACUGGCAGUUGACUCAUUUGCCGGGAUC ...----------------...(((((.....)).)))((----((((((((((((((.-.((.((((....(........)....)))).))..))))))))))))))))... ( -38.70) >DroYak_CAF1 36661 109 + 1 AAUGCAGAUAUAUACUCAAACAGACUUUAUUUCGAGUGCC----CGGCAAGUGGGUCGC-AUUGCAGUGAUGUAAUCAGCGGCAUAGCUGCCAGUUGACUCACUUGCCGGGAUC ............(((((................)))))((----(((((((((((((((-((((((....))))))..))((((....))))....)))))))))))))))... ( -46.39) >consensus AAUACACCACUGU__GCG_______AGUAUAUCGAGUGCCCAAGCGGCAAGUGGGUCGCCAGUGCAGUGAUGUAAUCGGCGGAAUAACUGGCAGUUGACUCACUUGCCGGGAUC ............................................((((((((((((((((..((((....))))...))).....(((.....))))))))))))))))..... (-26.54 = -28.96 + 2.42)


| Location | 8,921,661 – 8,921,768 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 74.93 |
| Mean single sequence MFE | -33.29 |
| Consensus MFE | -19.56 |
| Energy contribution | -19.76 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8921661 107 - 22224390 GAUUCCGGCAAGUGAGUCAACUGGCAGUUAUUUGGCCGAUUACAUCACUGCACUGGCGACCCACUUGCCGCUUGGGCACUCGAUAUACU-------CGCGUACACUGAUGCAUU ......((((((((.(((..(..(((((....((........))..)))))...)..))).))))))))(((..((.((.(((.....)-------)).)).).)..).))... ( -34.10) >DroSec_CAF1 35654 97 - 1 GAUCCCGGCAAGUGAGUCAACUGCCAG--------CCGAUUACAUCACUGCACUGGCGACCCACUUGCCGCUUGGCCACUCGAUAUACU-------CGC--ACAGUGGUGUAUU .....(((((((((.(((....(((((--------.................)))))))).)))))))))....(((((((((.....)-------)).--..))))))..... ( -33.43) >DroSim_CAF1 31993 105 - 1 GAUCCCGGCAAGUGAGUCAACUGCCAGCUAUUCCGCCGAUUACAUCACUGCACUGGCGACCCACUUGCCGCUUGGCCACUCGAUAUACU-------CGC--AAAGUGGUGUAUU .....(((((((((.(((....(((((.........................)))))))).)))))))))....(((((((((.....)-------)).--..))))))..... ( -33.01) >DroEre_CAF1 35856 93 - 1 GAUCCCGGCAAAUGAGUCAACUGCCAGUUAUUCCGUUGAUUGCAUCACUGCCAU-GCGACCCACUUGCCG----GGCACUCAACAUACGGUGUGU----------------AUU ....(((((((.((.(((...(((.(((((......)))))))).....((...-))))).)).))))))----)(((((........)))))..----------------... ( -26.50) >DroYak_CAF1 36661 109 - 1 GAUCCCGGCAAGUGAGUCAACUGGCAGCUAUGCCGCUGAUUACAUCACUGCAAU-GCGACCCACUUGCCG----GGCACUCGAAAUAAAGUCUGUUUGAGUAUAUAUCUGCAUU ...(((((((((((.(((....((((....))))((((((...))))..))...-..))).)))))))))----)).(((((((..........)))))))............. ( -39.40) >consensus GAUCCCGGCAAGUGAGUCAACUGCCAGCUAUUCCGCCGAUUACAUCACUGCACUGGCGACCCACUUGCCGCUUGGGCACUCGAUAUACU_______CGC__ACAGUGGUGCAUU .....(((((((((.(((...................(((...)))...((....))))).)))))))))............................................ (-19.56 = -19.76 + 0.20)

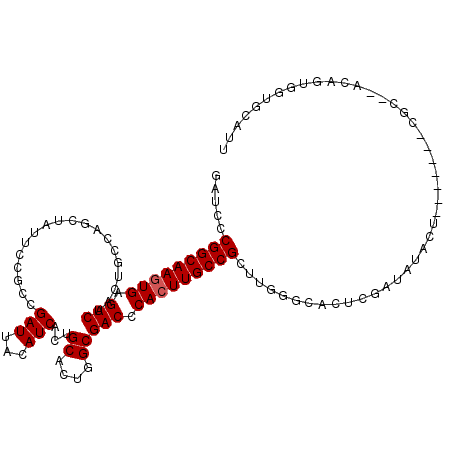
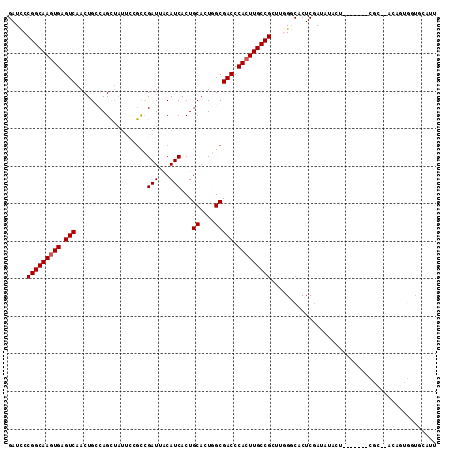
| Location | 8,921,693 – 8,921,808 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 87.16 |
| Mean single sequence MFE | -42.08 |
| Consensus MFE | -29.92 |
| Energy contribution | -32.66 |
| Covariance contribution | 2.74 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8921693 115 + 22224390 CCAAGCGGCAAGUGGGUCGCCAGUGCAGUGAUGUAAUCGGCCAAAUAACUGCCAGUUGACUCACUUGCCGGAAUCCGAGCUCCGAGAUCACUCAAUAGUUCGCACAUUGCCCAAU .....((((((((((((((((..((((....))))...))).....(((.....)))))))))))))))).....((((((..(((....)))...))))))............. ( -41.10) >DroSec_CAF1 35684 107 + 1 CCAAGCGGCAAGUGGGUCGCCAGUGCAGUGAUGUAAUCGG--------CUGGCAGUUGACUCACUUGCCGGGAUCCGAGCUCCAAGAUCACUCAAUAGCUCGCACAUUGCCCAAU ......(((((((((((((((((((((....))).....)--------)))))....))))))))))))(((...((((((....((....))...)))))).......)))... ( -44.00) >DroSim_CAF1 32023 115 + 1 CCAAGCGGCAAGUGGGUCGCCAGUGCAGUGAUGUAAUCGGCGGAAUAGCUGGCAGUUGACUCACUUGCCGGGAUCCGAGCUCCAAGAUCACUCAAUAGCUCGCACAUUGCCCAAU ......(((((((((((((((((((((....)))..((....))...))))))....))))))))))))(((...((((((....((....))...)))))).......)))... ( -44.30) >DroEre_CAF1 35879 110 + 1 C----CGGCAAGUGGGUCGC-AUGGCAGUGAUGCAAUCAACGGAAUAACUGGCAGUUGACUCAUUUGCCGGGAUCCGUGCUAUAAGAUCACUCAAUAACUCGCACAUUGCUCAAU (----((((((((((((((.-.((.((((....(........)....)))).))..))))))))))))))).....((((.....................)))).......... ( -37.60) >DroYak_CAF1 36700 110 + 1 C----CGGCAAGUGGGUCGC-AUUGCAGUGAUGUAAUCAGCGGCAUAGCUGCCAGUUGACUCACUUGCCGGGAUCCGAGCUCCAAGAUCACUCAAUAACUCGCACAUUGCCUAAU (----(((((((((((((((-((((((....))))))..))((((....))))....))))))))))))))((((..........))))............((.....))..... ( -43.40) >consensus CCAAGCGGCAAGUGGGUCGCCAGUGCAGUGAUGUAAUCGGCGGAAUAACUGGCAGUUGACUCACUUGCCGGGAUCCGAGCUCCAAGAUCACUCAAUAGCUCGCACAUUGCCCAAU ......(((((((((((((((..((((....))))...))).....(((.....)))))))))))))))(((...((((((....((....))...)))))).......)))... (-29.92 = -32.66 + 2.74)



| Location | 8,921,768 – 8,921,870 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.37 |
| Mean single sequence MFE | -31.45 |
| Consensus MFE | -18.30 |
| Energy contribution | -19.42 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8921768 102 - 22224390 CUUAU----------UGCUCUCAAGUGAAUGGGUGUGUU--------UUCGAGGCAUAUAGUAUUUUGUACAUGAUCAACAUUGGGCAAUGUGCGAACUAUUGAGUGAUCUCGGAGCUCG .....----------.((.((((......)))).))..(--------((((((((((.(((((.(((((((((..(((....)))...))))))))).))))).))).)))))))).... ( -28.20) >DroSec_CAF1 35751 120 - 1 UUUAUUUUCACCACUUGCUCUGAAGUGGAUGGGUGUGUUUUUUUUUUUUUGAGGCAUAUAGUAUUUUGUUCAUGAUCAGCAUUGGGCAAUGUGCGAGCUAUUGAGUGAUCUUGGAGCUCG .......(((((((((......)))))).)))(((((((((.........))))))))).((((.(((((((((.....)).))))))).))))(((((.(..((....))..)))))). ( -34.40) >DroSim_CAF1 32098 118 - 1 CUUAUUUUCACCACUUGCUCUCAAGUGGAUGGGUGUGUUUUU--UUUUUUGAGGCAUAUAGUAUUUUGUUCGUGAUCAGCAUUGGGCAAUGUGCGAGCUAUUGAGUGAUCUUGGAGCUCG .......((((((((((....))))))).)))(((((((((.--......))))))))).((((.(((((((((.....)).))))))).))))(((((.(..((....))..)))))). ( -36.20) >DroEre_CAF1 35949 107 - 1 CCCGUUU-CCCCAAUUUCUCUCAAUGGGAUAGGUGCUU------------UCGGCACGCAGUAUUCUGUACAUGAUCAGCAUUGAGCAAUGUGCGAGUUAUUGAGUGAUCUUAUAGCACG ((((((.-..............))))))....(((((.------------..(((((.((((((((.((((((..((......))...))))))))).))))).))).))....))))). ( -28.76) >DroYak_CAF1 36770 108 - 1 CUCGUUUUCACCACUUGCUCUCAAGUGGCUGGGUGUUU------------UAGGCAUAUAGUAUUUUGUACGUGAUCAGCAUUAGGCAAUGUGCGAGUUAUUGAGUGAUCUUGGAGCUCG .......((((((((((....))))))).)))(.((((------------(((((((.(((((.(((((((((..((.......))..))))))))).))))).)))..)))))))).). ( -29.70) >consensus CUUAUUUUCACCACUUGCUCUCAAGUGGAUGGGUGUGUU________UU_GAGGCAUAUAGUAUUUUGUACAUGAUCAGCAUUGGGCAAUGUGCGAGCUAUUGAGUGAUCUUGGAGCUCG ..........(((((((....))))))).........................((((((.((....(((.........)))....)).))))))(((((.(((((....)))))))))). (-18.30 = -19.42 + 1.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:50 2006