
| Sequence ID | X_DroMel_CAF1 |
|---|---|
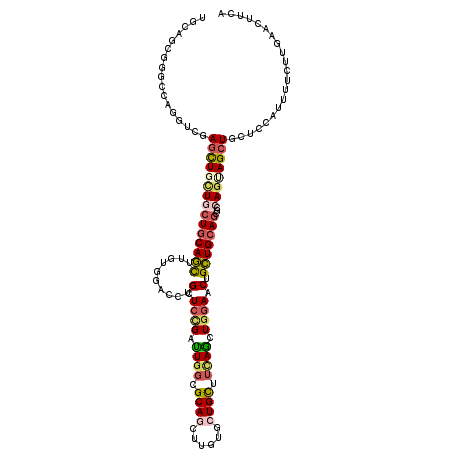
| Location | 8,901,366 – 8,901,486 |
| Length | 120 |
| Max. P | 0.778162 |

| Location | 8,901,366 – 8,901,486 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.06 |
| Mean single sequence MFE | -46.08 |
| Consensus MFE | -23.29 |
| Energy contribution | -23.93 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8901366 120 + 22224390 AGUCUCUGGCCAAAUCGAGCUGCUGCUGCAGCCGAUUGACCUGCUCCGAUUGGCGCAGCUUGUGCUGCUUCAGCUGGAACUGCUGCAGUCCGAGUAGCUGCUCCAUCUUGCGCAGCUUCA .......((...(((((.(((((....))))))))))..))(((((.(((((..(((((....)))))..((((.......))))))))).)))))(((((.(......).))))).... ( -48.60) >DroVir_CAF1 22079 120 + 1 UGCAGCGCGUCAGAUCGAGCUGCUGCUGCAGCUUGUGCAGCUGCUCGGAUUGGCGCAGAUUGUGCUGCUUGAGCUGGAACUGCUGCAGGGCCAGCAGCUGCUCCAUUUUCUUGAACUUGA .((((((((((((.((((((.(((((.((.....))))))).)))))).)))))))................(((((..(((...)))..))))).)))))................... ( -54.20) >DroGri_CAF1 18046 120 + 1 UACAUCUGGUCAGGUCGAGUUGCUGCUGCAACUUGUGCAACUGCUCCGAUUGGCGCAAAUUGUGCUGUUUUAGCUGAAACUGUUGCAACGCCAGUAGCUGCUCCAAUUUCUUGAACUUCA .........(((((..((((.(((((((.......((((((..........(((((.....)))))(((((....))))).))))))....))))))).))))......)))))...... ( -34.20) >DroWil_CAF1 23258 120 + 1 UGCUGCGAGCGUGUUCCAACUGCUGCUGCAGCUUCUGUACCUGCUCCGAUUGCCGCAGCUUAUGCUGCUUUAAUUGGAACUGCUGAAGAGCCAAUAGCUGUUCCAUUUUCUUAAACUUCA .(((((.((((.((.......)))))))))))..........((((((((((..(((((....)))))..))))))))...)).(((.(((.....))).)))................. ( -36.40) >DroMoj_CAF1 22543 120 + 1 UGGUGCGCGUCAGGUCGAGCUGUUGCUGCAGUUUCUGGAGCUGCUCCGACUGGCGCAAGUUGUGCUGCUUCAGCUGGAACUGCUGCAGGGCGAGGAGCUGCUCCAUUUUCUUGAACUUCA .........(((((..(((((((....))))))).((((((.(((((....(((....)))((.((((..(((......)))..)))).))..))))).))))))....)))))...... ( -47.00) >DroAna_CAF1 15729 120 + 1 UGCAGUGGGCGUGGUCCAGUUGCUGCUGCAGUCGGCCGACGUGCUCUGACUGGCGCAGCUUGUGCUGCUUCAGCUGGAACUGCUGCAGCCCCAGGAGCUGCUCCAUCUUGCGCAGCUUCA (((((..(((((((.((..((((....))))..)))).))))..(((..((((.(((((....))))).))))..))).)..)))))......((((((((.(......).)))))))). ( -56.10) >consensus UGCAGCGGGCCAGGUCGAGCUGCUGCUGCAGCUUGUGGACCUGCUCCGAUUGGCGCAGCUUGUGCUGCUUCAGCUGGAACUGCUGCAGCGCCAGUAGCUGCUCCAUUUUCUUGAACUUCA .................(((((((((((((((..........(.((((.((((.((((......)))).)))).)))).).)))))))...))))))))..................... (-23.29 = -23.93 + 0.64)

| Location | 8,901,366 – 8,901,486 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.06 |
| Mean single sequence MFE | -42.24 |
| Consensus MFE | -21.61 |
| Energy contribution | -21.78 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8901366 120 - 22224390 UGAAGCUGCGCAAGAUGGAGCAGCUACUCGGACUGCAGCAGUUCCAGCUGAAGCAGCACAAGCUGCGCCAAUCGGAGCAGGUCAAUCGGCUGCAGCAGCAGCUCGAUUUGGCCAGAGACU ....(((.((.....(((.((((((....(..((((..((((....))))..))))..).)))))).)))..)).))).((((((((((((((....))))).))).))))))....... ( -51.40) >DroVir_CAF1 22079 120 - 1 UCAAGUUCAAGAAAAUGGAGCAGCUGCUGGCCCUGCAGCAGUUCCAGCUCAAGCAGCACAAUCUGCGCCAAUCCGAGCAGCUGCACAAGCUGCAGCAGCAGCUCGAUCUGACGCGCUGCA ....((((..((...((((((.(((((.......))))).))))))..))..((((......))))........)))).((((((.....)))))).(((((.((......)).))))). ( -48.60) >DroGri_CAF1 18046 120 - 1 UGAAGUUCAAGAAAUUGGAGCAGCUACUGGCGUUGCAACAGUUUCAGCUAAAACAGCACAAUUUGCGCCAAUCGGAGCAGUUGCACAAGUUGCAGCAGCAACUCGACCUGACCAGAUGUA .....(((((....)))))(((....(((((..((((((.......(((.....)))......(((.((....)).)))))))))(.((((((....)))))).)....).)))).))). ( -32.90) >DroWil_CAF1 23258 120 - 1 UGAAGUUUAAGAAAAUGGAACAGCUAUUGGCUCUUCAGCAGUUCCAAUUAAAGCAGCAUAAGCUGCGGCAAUCGGAGCAGGUACAGAAGCUGCAGCAGCAGUUGGAACACGCUCGCAGCA (((((....((..(((((.....)))))..)))))))...(((((.(((...(((((....)))))...))).)))))..........((((((((..............))).))))). ( -35.14) >DroMoj_CAF1 22543 120 - 1 UGAAGUUCAAGAAAAUGGAGCAGCUCCUCGCCCUGCAGCAGUUCCAGCUGAAGCAGCACAACUUGCGCCAGUCGGAGCAGCUCCAGAAACUGCAGCAACAGCUCGACCUGACGCGCACCA ...............((((((.(((((......(((..((((....))))..)))(((.....))).......))))).)))))).....(((.((..(((......)))..)))))... ( -36.60) >DroAna_CAF1 15729 120 - 1 UGAAGCUGCGCAAGAUGGAGCAGCUCCUGGGGCUGCAGCAGUUCCAGCUGAAGCAGCACAAGCUGCGCCAGUCAGAGCACGUCGGCCGACUGCAGCAGCAACUGGACCACGCCCACUGCA ....(((((((..(((((.((((((..((..(((((..((((....))))..))))).)))))))).))).))...))..(((....))).))))).(((..(((.......))).))). ( -48.80) >consensus UGAAGUUCAAGAAAAUGGAGCAGCUACUGGCCCUGCAGCAGUUCCAGCUGAAGCAGCACAAGCUGCGCCAAUCGGAGCAGGUCCACAAGCUGCAGCAGCAGCUCGACCUGACCCGCUGCA ....((((.......((((((.(((.(.......).))).))))))......((((......))))........)))).........((((((....))))))................. (-21.61 = -21.78 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:44 2006