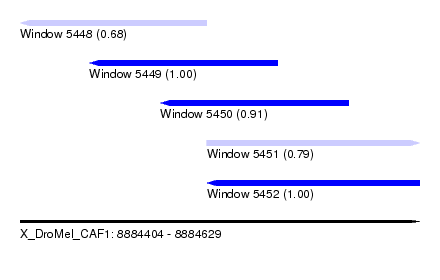
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,884,404 – 8,884,629 |
| Length | 225 |
| Max. P | 0.999587 |

| Location | 8,884,404 – 8,884,509 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.52 |
| Mean single sequence MFE | -29.47 |
| Consensus MFE | -16.85 |
| Energy contribution | -17.60 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8884404 105 - 22224390 CGGCUAAAAAGA---------AAAAGGUUGG-----AAAAACGUUCGACGUUUUGGCCAAGACUGAAAAACAAGUGCCAACUUGAGGAUGAUUCAAGGCGCGUUAAUGCAUC-AGCCAGC .((((.......---------......((((-----.((((((.....))))))..))))((.((........(((((...(((((.....)))))))))).......))))-))))... ( -26.56) >DroSec_CAF1 14928 112 - 1 CGGCAAAAAA--AAAAAGAAAAAAAGGUUGG-----AAAAACGUCCGGCGUUUUGGCCAAGACUGAAAAACAAGUGCCAACUUGAGGAUGAUUCAAGGCGCGUUAAUGCAUC-GGCCAGC ..((......--.........((((.(((((-----(......)))))).))))((((...............(((((...(((((.....))))))))))((....))...-)))).)) ( -28.80) >DroSim_CAF1 15063 114 - 1 CGGCAAAAAAAAAAAAAGGAAAAAAUGUUGG-----AAAAACGUCCGGCGUUUUGGCCAAGACUGAAAAACAAGUGCCAACUUGAGGAUGAUUCAAGGCGCGUUAAUGCAUC-GGCCAAC .(((.............((..((((((((((-----(......)))))))))))..))..((.((........(((((...(((((.....)))))))))).......))))-.)))... ( -32.46) >DroEre_CAF1 13529 111 - 1 CGGCAAAAAAAA---AAGAAGUAAUUGUUGG-----AAAAACGUCCGGCGUUUUGGCCAAGACUGAAAAACAAGUGCCAACUUGAGGAUGAUUCAAGGCGCGUUAAUGCAUC-AGCCAGC .(((........---..........(.((((-----.((((((.....))))))..)))).).(((.......(((((...(((((.....))))))))))((....)).))-))))... ( -26.30) >DroYak_CAF1 13924 111 - 1 CGGCAAAAAAAA---AAAGAAAAAAGCUUGG-----AAAAACGUCGGGCGUUUUGGCCAAGACUGAAAAACAAGUGCCAACUUGAGGAUGAUUCAAGGCGUGUUAAUGCAUC-AGCCAGC .(((........---...(((...(((((((-----.((((((.....))))))..))))).))......(((((....))))).......)))...((((....))))...-.)))... ( -28.90) >DroAna_CAF1 17012 97 - 1 CGGCAGAGCGGA--------AAAUGGCUUCGCGGCUGAAAACG-----------GGCCAGG--UG-AGCCCAGGUGCCGGCCUGCGGAUGAUUCAAGGCGCUGUCG-GAGUCUACCCAGG ((((((..(((.--------...(((.(((((((((.......-----------))))..)--))-)).)))....)))((((...((....)).)))).))))))-............. ( -33.80) >consensus CGGCAAAAAAAA___AAG_AAAAAAGGUUGG_____AAAAACGUCCGGCGUUUUGGCCAAGACUGAAAAACAAGUGCCAACUUGAGGAUGAUUCAAGGCGCGUUAAUGCAUC_AGCCAGC .(((.................................((((((.....)))))).(((............(((((....)))))..((....))..)))...............)))... (-16.85 = -17.60 + 0.75)

| Location | 8,884,443 – 8,884,549 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.43 |
| Mean single sequence MFE | -24.72 |
| Consensus MFE | -20.38 |
| Energy contribution | -20.62 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.82 |
| SVM decision value | 3.75 |
| SVM RNA-class probability | 0.999587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8884443 106 - 22224390 AAAAAAAAAAAAACAACAAAACUAUGGCUUCGCCAACUUCCGGCUAAAAAGA---------AAAAGGUUGGAAAAACGUUCGACGUUUUGGCCAAGACUGAAAAACAAGUGCCAA ........................((((...((((..((((((((.......---------....))))))))(((((.....)))))))))....(((........))))))). ( -22.50) >DroSim_CAF1 15102 97 - 1 ------------------GCACUAUGGCUGCGCCAACUUCCGGCAAAAAAAAAAAAAGGAAAAAAUGUUGGAAAAACGUCCGGCGUUUUGGCCAAGACUGAAAAACAAGUGCCAA ------------------(((((.((.....(((.......))).............((..(((((((((((......)))))))))))..))............)))))))... ( -27.30) >DroEre_CAF1 13568 94 - 1 ------------------AGACUAUGGCUUCGCCAACUUCCGGCAAAAAAAA---AAGAAGUAAUUGUUGGAAAAACGUCCGGCGUUUUGGCCAAGACUGAAAAACAAGUGCCAA ------------------......((((...((((..(((((((((......---.........)))))))))(((((.....)))))))))....(((........))))))). ( -23.16) >DroYak_CAF1 13963 94 - 1 ------------------AGACUAUGGCUUGGCCAACUUCCGGCAAAAAAAA---AAAGAAAAAAGCUUGGAAAAACGUCGGGCGUUUUGGCCAAGACUGAAAAACAAGUGCCAA ------------------......(((((((((((((.((((((........---.(((.......)))........)))))).))..))))))).(((........))))))). ( -25.93) >consensus __________________AGACUAUGGCUUCGCCAACUUCCGGCAAAAAAAA___AAGGAAAAAAGGUUGGAAAAACGUCCGGCGUUUUGGCCAAGACUGAAAAACAAGUGCCAA ........................((((...((((..((((((((....................))))))))(((((.....)))))))))....(((........))))))). (-20.38 = -20.62 + 0.25)


| Location | 8,884,483 – 8,884,589 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.47 |
| Mean single sequence MFE | -20.06 |
| Consensus MFE | -15.26 |
| Energy contribution | -15.13 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8884483 106 - 22224390 GGCAUGCCCAUAUACGAUCACGAAUUGGCCAGGUUAAAAAAAAAAAAAAAAAACAACAAAACUAUGGCUUCGCCAACUUCCGGCUAAAAAGA---------AAAAGGUUGGAAAA (((...........((....))....(((((((((........................)))).)))))..)))...((((((((.......---------....)))))))).. ( -18.46) >DroSim_CAF1 15142 97 - 1 GGCAUGCCCAUAUACGAUCACGAAUUGGCCAGGUCCAAAA------------------GCACUAUGGCUGCGCCAACUUCCGGCAAAAAAAAAAAAAGGAAAAAAUGUUGGAAAA (((...........((....))....((((((((.(....------------------).))).)))))..)))...((((((((....................)))))))).. ( -19.05) >DroEre_CAF1 13608 94 - 1 GGCAUGCCCAUAUACGAUCACGAAUUUGCCAGGUCCAAAA------------------AGACUAUGGCUUCGCCAACUUCCGGCAAAAAAAA---AAGAAGUAAUUGUUGGAAAA (((...........((....)).....((((((((.....------------------.)))).))))...)))...(((((((((......---.........))))))))).. ( -22.16) >DroYak_CAF1 14003 94 - 1 GGUAUGCCCAUAUACGAUCACGAAUUGGUCAGGUCCAAAA------------------AGACUAUGGCUUGGCCAACUUCCGGCAAAAAAAA---AAAGAAAAAAGCUUGGAAAA .(((((....))))).........(((((((((.(((...------------------......)))))))))))).(((((((........---..........)).))))).. ( -20.57) >consensus GGCAUGCCCAUAUACGAUCACGAAUUGGCCAGGUCCAAAA__________________AGACUAUGGCUUCGCCAACUUCCGGCAAAAAAAA___AAGGAAAAAAGGUUGGAAAA (((...........((....)).....((((((((........................)))).))))...)))...((((((((....................)))))))).. (-15.26 = -15.13 + -0.13)


| Location | 8,884,509 – 8,884,629 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.55 |
| Mean single sequence MFE | -30.46 |
| Consensus MFE | -24.75 |
| Energy contribution | -25.19 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8884509 120 + 22224390 GAAGUUGGCGAAGCCAUAGUUUUGUUGUUUUUUUUUUUUUUUUUUAACCUGGCCAAUUCGUGAUCGUAUAUGGGCAUGCCAUCACGUAGUUAAUAGAAAGUGGACAGGCCCAAAAGCGUA (((.((((((((((....)))))((((.................))))...)))))))).....(((.(.(((((.((((((.................)))).)).))))).).))).. ( -24.06) >DroSec_CAF1 15040 102 + 1 GAAGUUGGCAAAGCCAUAGUGC------------------UUUUGGACCUGGCCAAUUCGUGAUCGUAUAUGGGCAUGCCAUCACGUAGUUAAUAGAAAGUGGACAGGCCCAAAAGCGAA .....((((...))))...(((------------------((((((.((((.(((...((((((.((((......)))).))))))..............))).)))).))))))))).. ( -34.63) >DroSim_CAF1 15177 102 + 1 GAAGUUGGCGCAGCCAUAGUGC------------------UUUUGGACCUGGCCAAUUCGUGAUCGUAUAUGGGCAUGCCAUCACGUAGUUAAUAGAAAGUGGACAGGCCCAAAAGCGAA .....((((...))))...(((------------------((((((.((((.(((...((((((.((((......)))).))))))..............))).)))).))))))))).. ( -34.63) >DroEre_CAF1 13640 102 + 1 GAAGUUGGCGAAGCCAUAGUCU------------------UUUUGGACCUGGCAAAUUCGUGAUCGUAUAUGGGCAUGCCAUCACGUAGUUAAUAGAAAGUGGACAGGCCCAAAAGCGAA .....(.(((((((((..((((------------------....)))).))))...))))).)((((.(.(((((.((((((.................)))).)).))))).).)))). ( -29.73) >DroYak_CAF1 14035 102 + 1 GAAGUUGGCCAAGCCAUAGUCU------------------UUUUGGACCUGACCAAUUCGUGAUCGUAUAUGGGCAUACCAUCACGUAGUUAAUAGAAAGUGGACAGGCCCAAAAGCAAA ((...((((...))))...))(------------------((((((.((((.(((...((((((.((((......)))).))))))..............))).)))).))))))).... ( -29.23) >consensus GAAGUUGGCGAAGCCAUAGUCU__________________UUUUGGACCUGGCCAAUUCGUGAUCGUAUAUGGGCAUGCCAUCACGUAGUUAAUAGAAAGUGGACAGGCCCAAAAGCGAA .....((((...))))........................((((((.((((.(((...((((((.((((......)))).))))))..............))).)))).))))))..... (-24.75 = -25.19 + 0.44)



| Location | 8,884,509 – 8,884,629 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.55 |
| Mean single sequence MFE | -31.53 |
| Consensus MFE | -26.08 |
| Energy contribution | -26.32 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.50 |
| SVM RNA-class probability | 0.999304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8884509 120 - 22224390 UACGCUUUUGGGCCUGUCCACUUUCUAUUAACUACGUGAUGGCAUGCCCAUAUACGAUCACGAAUUGGCCAGGUUAAAAAAAAAAAAAAAAAACAACAAAACUAUGGCUUCGCCAACUUC ..((..(.(((((.(((((((..............)))..)))).))))).)..)).....((((((((.((((((............................)))))).))))).))) ( -23.73) >DroSec_CAF1 15040 102 - 1 UUCGCUUUUGGGCCUGUCCACUUUCUAUUAACUACGUGAUGGCAUGCCCAUAUACGAUCACGAAUUGGCCAGGUCCAAAA------------------GCACUAUGGCUUUGCCAACUUC ...(((((((((((((.(((.((...........((((((.(.(((....))).).)))))))).))).)))))))))))------------------))....((((...))))..... ( -36.20) >DroSim_CAF1 15177 102 - 1 UUCGCUUUUGGGCCUGUCCACUUUCUAUUAACUACGUGAUGGCAUGCCCAUAUACGAUCACGAAUUGGCCAGGUCCAAAA------------------GCACUAUGGCUGCGCCAACUUC ...(((((((((((((.(((.((...........((((((.(.(((....))).).)))))))).))).)))))))))))------------------))....((((...))))..... ( -36.20) >DroEre_CAF1 13640 102 - 1 UUCGCUUUUGGGCCUGUCCACUUUCUAUUAACUACGUGAUGGCAUGCCCAUAUACGAUCACGAAUUUGCCAGGUCCAAAA------------------AGACUAUGGCUUCGCCAACUUC .(((..(.(((((.(((((((..............)))..)))).))))).)..)))...((((...((((((((.....------------------.)))).))))))))........ ( -26.74) >DroYak_CAF1 14035 102 - 1 UUUGCUUUUGGGCCUGUCCACUUUCUAUUAACUACGUGAUGGUAUGCCCAUAUACGAUCACGAAUUGGUCAGGUCCAAAA------------------AGACUAUGGCUUGGCCAACUUC .....(((((((((((.(((.((...........((((((.(((((....))))).)))))))).))).)))))))))))------------------......((((...))))..... ( -34.80) >consensus UUCGCUUUUGGGCCUGUCCACUUUCUAUUAACUACGUGAUGGCAUGCCCAUAUACGAUCACGAAUUGGCCAGGUCCAAAA__________________ACACUAUGGCUUCGCCAACUUC .....(((((((((((.(((.((...........((((((.(.(((....))).).)))))))).))).)))))))))))........................((((...))))..... (-26.08 = -26.32 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:39 2006