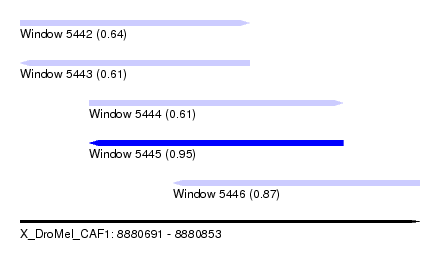
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,880,691 – 8,880,853 |
| Length | 162 |
| Max. P | 0.953552 |

| Location | 8,880,691 – 8,880,784 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.73 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -17.75 |
| Energy contribution | -18.64 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8880691 93 + 22224390 ----------G-GAACGGGCGAAGAACUGUUCUCAUUUC-------UCAUUUCGCGACGGUUGGAAAGUAUUCAAAUGCCUCA-AUGGAAACA----UUCCCUUUCUCCGCCA----ACU ----------(-(..(((((((((((.((....)).)))-------)....))))((.(((..(((....)))....)))))(-(((....))----))........))))).----... ( -19.70) >DroSec_CAF1 11201 94 + 1 ----------G-GAACGGGCGAAGAACUGUUCUCAUUUC-------UCAUUUCGCGGCGGUUGGAAAGUAUUCAAAUGCCUCAAAUGGAGACA----UUCCCUUUCUCCGCCA----UUU ----------(-((((((........)))))))......-------.........(((((..((((((......((((.(((.....))).))----))..))))))))))).----... ( -26.00) >DroSim_CAF1 11338 94 + 1 ----------G-GAACGGGCGAAGAACUGUUCUCAUUUC-------UCAUUUCGCGGCGGUUGGAAAGUAUUCAAAUGCCUCAAAUGGAGACA----UUCCCUUUCUCCGCCA----UCU ----------(-((((((........)))))))......-------.........(((((..((((((......((((.(((.....))).))----))..))))))))))).----... ( -26.00) >DroEre_CAF1 10088 94 + 1 ----------GCGAACGGACGGAGAACUGUUCUCAUUUC-------GCAUUUCACGGCGGUUGGAAAGUAUUCAAAUGCCUCA-ACGGAGACA----UUCCCUUUCUCCGCCA----UCU ----------(((((.((((((....))))))....)))-------)).......(((((..((((((......((((.(((.-...))).))----))..))))))))))).----... ( -29.30) >DroYak_CAF1 11087 100 + 1 ----------G-GAACGCGCGGAGAACUGUUCUCAUUUCUCAUUUCUCAUUUCUCGACGGUUGGAAAGUAUUCAAAUGCCUCA-AUGGAGACA----UUCCCUUUCUCCGUCA----UCU ----------(-(((.(.(..(((((.((....)).)))))....).).))))..(((((..((((((......((((.(((.-...))).))----))..))))))))))).----... ( -22.40) >DroAna_CAF1 14779 102 + 1 CCGAUCUCGGG-GAAAAGGC--AGAAACGUUCUCGUUUC--------------UCGCCAGUUGGAAAGUAUUCAAAUGCCUCA-AUGGAGUCGCUGUUUUCCUUUCUCCGCUCCACAAUU .......((((-((((.(((--(((((((....))))))--------------).)))....(((((((((....))))(((.-...))).......))))))))))))).......... ( -29.60) >consensus __________G_GAACGGGCGAAGAACUGUUCUCAUUUC_______UCAUUUCGCGGCGGUUGGAAAGUAUUCAAAUGCCUCA_AUGGAGACA____UUCCCUUUCUCCGCCA____UCU ............((((((........)))))).......................(((((..((((((........((.(((.....))).))........)))))))))))........ (-17.75 = -18.64 + 0.89)


| Location | 8,880,691 – 8,880,784 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.73 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -18.10 |
| Energy contribution | -17.94 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8880691 93 - 22224390 AGU----UGGCGGAGAAAGGGAA----UGUUUCCAU-UGAGGCAUUUGAAUACUUUCCAACCGUCGCGAAAUGA-------GAAAUGAGAACAGUUCUUCGCCCGUUC-C---------- ...----.(((((.(((((.(((----((((((...-.))))))))).....)))))...)))))((((....(-------(((.((....)).))))))))......-.---------- ( -24.00) >DroSec_CAF1 11201 94 - 1 AAA----UGGCGGAGAAAGGGAA----UGUCUCCAUUUGAGGCAUUUGAAUACUUUCCAACCGCCGCGAAAUGA-------GAAAUGAGAACAGUUCUUCGCCCGUUC-C---------- ...----.(((((.(((((.(((----((((((.....))))))))).....)))))...)))))((((....(-------(((.((....)).))))))))......-.---------- ( -29.20) >DroSim_CAF1 11338 94 - 1 AGA----UGGCGGAGAAAGGGAA----UGUCUCCAUUUGAGGCAUUUGAAUACUUUCCAACCGCCGCGAAAUGA-------GAAAUGAGAACAGUUCUUCGCCCGUUC-C---------- ...----.(((((.(((((.(((----((((((.....))))))))).....)))))...)))))((((....(-------(((.((....)).))))))))......-.---------- ( -29.20) >DroEre_CAF1 10088 94 - 1 AGA----UGGCGGAGAAAGGGAA----UGUCUCCGU-UGAGGCAUUUGAAUACUUUCCAACCGCCGUGAAAUGC-------GAAAUGAGAACAGUUCUCCGUCCGUUCGC---------- ..(----((((((.(((((.(((----((((((...-.))))))))).....)))))...))))))).....((-------(((..((((.....))))......)))))---------- ( -31.70) >DroYak_CAF1 11087 100 - 1 AGA----UGACGGAGAAAGGGAA----UGUCUCCAU-UGAGGCAUUUGAAUACUUUCCAACCGUCGAGAAAUGAGAAAUGAGAAAUGAGAACAGUUCUCCGCGCGUUC-C---------- ...----((((((.(((((.(((----((((((...-.))))))))).....)))))...))))))...((((.(....(((((.((....)).)))))..).)))).-.---------- ( -26.90) >DroAna_CAF1 14779 102 - 1 AAUUGUGGAGCGGAGAAAGGAAAACAGCGACUCCAU-UGAGGCAUUUGAAUACUUUCCAACUGGCGA--------------GAAACGAGAACGUUUCU--GCCUUUUC-CCCGAGAUCGG .......((.(((.(((((((((....(((..((..-...))...))).....)))))....(((.(--------------((((((....)))))))--))).))))-.)))...)).. ( -26.80) >consensus AGA____UGGCGGAGAAAGGGAA____UGUCUCCAU_UGAGGCAUUUGAAUACUUUCCAACCGCCGCGAAAUGA_______GAAAUGAGAACAGUUCUUCGCCCGUUC_C__________ ........(((((.(((((........((((((.....))))))........)))))...)))))................(((.((....)).)))....................... (-18.10 = -17.94 + -0.16)

| Location | 8,880,719 – 8,880,822 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 84.29 |
| Mean single sequence MFE | -23.46 |
| Consensus MFE | -16.02 |
| Energy contribution | -16.18 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8880719 103 + 22224390 ------UCAUUUCGCGACGGUUGGAAAGUAUUCAAAUGCCUCA-AUGGAAACAUUCCCUUUCUCCGCCAACUCGAAUUCUAAAAUGUGUCCGUUUUUUUUCAUACCCUCU-- ------.........((.(((((((((((((....))))...(-(((((.(((((....(((...........)))......))))).))))))..)))))).))).)).-- ( -16.10) >DroSec_CAF1 11229 102 + 1 ------UCAUUUCGCGGCGGUUGGAAAGUAUUCAAAUGCCUCAAAUGGAGACAUUCCCUUUCUCCGCCAUUUCACAUGCG-AAAUGAUUCCGUUU-UUUUCAUACCCUCG-- ------((((((((((((((..((((((......((((.(((.....))).))))..))))))))))).........)))-))))))........-..............-- ( -27.40) >DroSim_CAF1 11366 102 + 1 ------UCAUUUCGCGGCGGUUGGAAAGUAUUCAAAUGCCUCAAAUGGAGACAUUCCCUUUCUCCGCCAUCUCGAAUUCG-AAAUGAUUCCGUUU-CUUUCAUACCCUCU-- ------(((((((..(((((..((((((......((((.(((.....))).))))..))))))))))).....(....))-))))))........-..............-- ( -24.30) >DroEre_CAF1 10117 102 + 1 ------GCAUUUCACGGCGGUUGGAAAGUAUUCAAAUGCCUCA-ACGGAGACAUUCCCUUUCUCCGCCAUCUCGAACUCG-AAAUGAUUCGG-UU-UUGUCUUACCCUCGAU ------.((((((..(((((..((((((......((((.(((.-...))).))))..))))))))))).....(....))-)))))....((-(.-.......)))...... ( -24.90) >DroYak_CAF1 11116 109 + 1 CAUUUCUCAUUUCUCGACGGUUGGAAAGUAUUCAAAUGCCUCA-AUGGAGACAUUCCCUUUCUCCGUCAUCUCGAGUUCG-AAAUGAAUCGACUU-UCGUCAUAUCCUCGAU ...............(((((..((((((......((((.(((.-...))).))))..)))))))))))...(((((...(-(.((((..((....-.)))))).))))))). ( -24.60) >consensus ______UCAUUUCGCGGCGGUUGGAAAGUAUUCAAAUGCCUCA_AUGGAGACAUUCCCUUUCUCCGCCAUCUCGAAUUCG_AAAUGAUUCCGUUU_UUUUCAUACCCUCG__ ...............(((((..((((((......((((.(((.....))).))))..)))))))))))............................................ (-16.02 = -16.18 + 0.16)

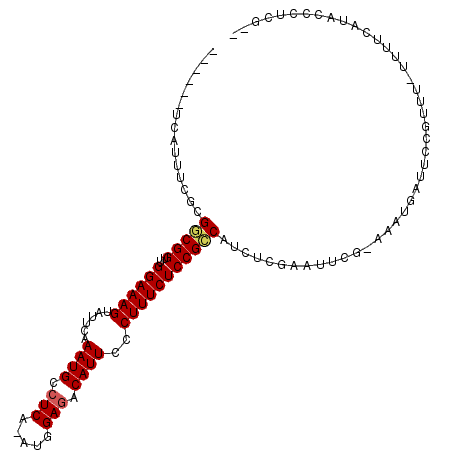
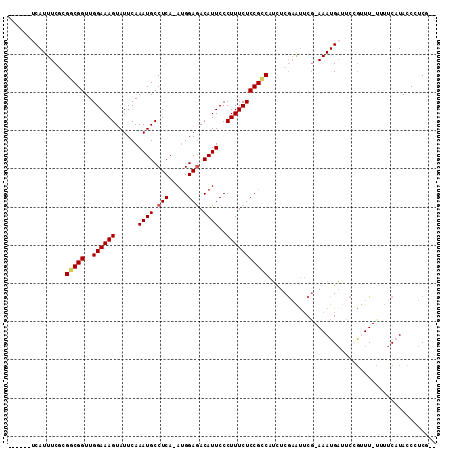
| Location | 8,880,719 – 8,880,822 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 84.29 |
| Mean single sequence MFE | -30.14 |
| Consensus MFE | -22.66 |
| Energy contribution | -22.14 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8880719 103 - 22224390 --AGAGGGUAUGAAAAAAAACGGACACAUUUUAGAAUUCGAGUUGGCGGAGAAAGGGAAUGUUUCCAU-UGAGGCAUUUGAAUACUUUCCAACCGUCGCGAAAUGA------ --........................((((((.((...((.(((((.((((.....(((((((((...-.))))))))).....)))))))))))))..)))))).------ ( -20.60) >DroSec_CAF1 11229 102 - 1 --CGAGGGUAUGAAAA-AAACGGAAUCAUUU-CGCAUGUGAAAUGGCGGAGAAAGGGAAUGUCUCCAUUUGAGGCAUUUGAAUACUUUCCAACCGCCGCGAAAUGA------ --..............-........((((((-(((((.....))(((((.(((((.(((((((((.....))))))))).....)))))...))))))))))))))------ ( -33.40) >DroSim_CAF1 11366 102 - 1 --AGAGGGUAUGAAAG-AAACGGAAUCAUUU-CGAAUUCGAGAUGGCGGAGAAAGGGAAUGUCUCCAUUUGAGGCAUUUGAAUACUUUCCAACCGCCGCGAAAUGA------ --..............-........((((((-((.........((((((.(((((.(((((((((.....))))))))).....)))))...))))))))))))))------ ( -29.60) >DroEre_CAF1 10117 102 - 1 AUCGAGGGUAAGACAA-AA-CCGAAUCAUUU-CGAGUUCGAGAUGGCGGAGAAAGGGAAUGUCUCCGU-UGAGGCAUUUGAAUACUUUCCAACCGCCGUGAAAUGC------ .((((((((.......-.)-))(((....))-)...))))).(((((((.(((((.(((((((((...-.))))))))).....)))))...))))))).......------ ( -32.10) >DroYak_CAF1 11116 109 - 1 AUCGAGGAUAUGACGA-AAGUCGAUUCAUUU-CGAACUCGAGAUGACGGAGAAAGGGAAUGUCUCCAU-UGAGGCAUUUGAAUACUUUCCAACCGUCGAGAAAUGAGAAAUG .((((((...((((..-..))))..((((((-((....)))))))).(((((....(((((((((...-.)))))))))......)))))..)).))))............. ( -35.00) >consensus __CGAGGGUAUGAAAA_AAACGGAAUCAUUU_CGAAUUCGAGAUGGCGGAGAAAGGGAAUGUCUCCAU_UGAGGCAUUUGAAUACUUUCCAACCGCCGCGAAAUGA______ ..........................(((((.((....))...((((((.(((((.(((((((((.....))))))))).....)))))...))))))..)))))....... (-22.66 = -22.14 + -0.52)

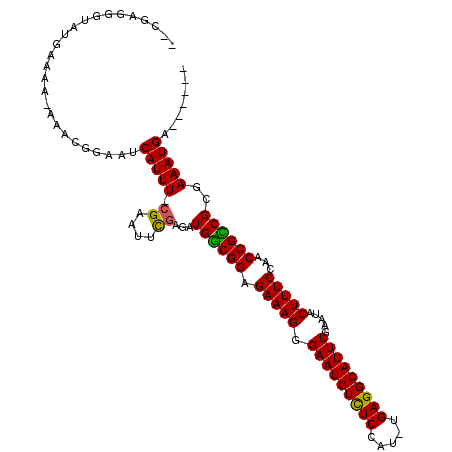
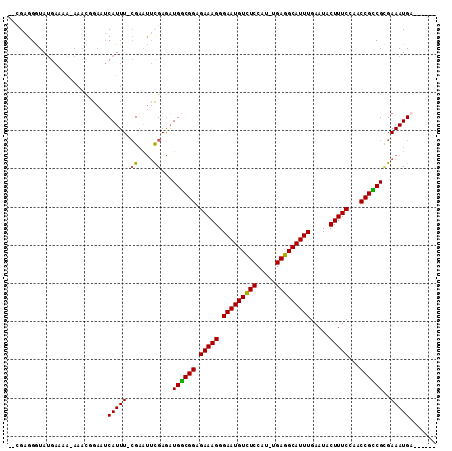
| Location | 8,880,753 – 8,880,853 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 72.98 |
| Mean single sequence MFE | -22.92 |
| Consensus MFE | -7.92 |
| Energy contribution | -7.84 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8880753 100 - 22224390 AAAGUGGAUAAAACUUUAGGGUAGCAACAAU-----------AGAGGGUAUGAAAAAAAACGGACACAUUUUAGAAUUCGAGUUGGCGGAGAAAGGGAAUGUUUCCAU-UGA .......(((...(((((..((....))..)-----------))))..)))..........(((.(((((((....((((......)))).....))))))).)))..-... ( -17.90) >DroSec_CAF1 11263 89 - 1 AAAA----------UAUAGGGUAGCAACUAU-----------CGAGGGUAUGAAAA-AAACGGAAUCAUUU-CGCAUGUGAAAUGGCGGAGAAAGGGAAUGUCUCCAUUUGA ....----------.....(((((...))))-----------).............-...((((.((((((-((....)))))))).(((((.........))))).)))). ( -15.70) >DroSim_CAF1 11400 89 - 1 AAAA----------UAUAGGGUAGCAACUAU-----------AGAGGGUAUGAAAG-AAACGGAAUCAUUU-CGAAUUCGAGAUGGCGGAGAAAGGGAAUGUCUCCAUUUGA ....----------(((((........))))-----------).............-...((((.((((((-((....)))))))).(((((.........))))).)))). ( -18.30) >DroEre_CAF1 10151 108 - 1 GAAAUGGAUAAGGCUGCAGGGUAUUAACAGUAGUUUAUCCAUCGAGGGUAAGACAA-AA-CCGAAUCAUUU-CGAGUUCGAGAUGGCGGAGAAAGGGAAUGUCUCCGU-UGA ....((((((((.((((.(........).))))))))))))((((.((..(((((.-..-((...((((((-((....))))))))........))...))))))).)-))) ( -31.70) >DroYak_CAF1 11156 109 - 1 AAAAUGCUUAAAGCUAUAGGGUAGCAACUAUAGUUUACACAUCGAGGAUAUGACGA-AAGUCGAUUCAUUU-CGAACUCGAGAUGACGGAGAAAGGGAAUGUCUCCAU-UGA ..........(((((((((........))))))))).....((((.....((((..-..))))..((((((-((....)))))))).(((((.........))))).)-))) ( -31.00) >consensus AAAAUG__UAA__CUAUAGGGUAGCAACUAU___________CGAGGGUAUGAAAA_AAACGGAAUCAUUU_CGAAUUCGAGAUGGCGGAGAAAGGGAAUGUCUCCAU_UGA ..................................................................(((((.((....)))))))..(((((.........)))))...... ( -7.92 = -7.84 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:33 2006