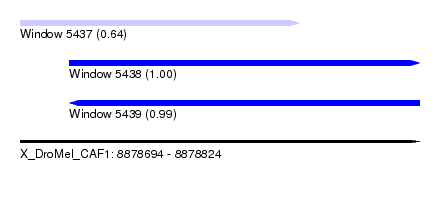
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,878,694 – 8,878,824 |
| Length | 130 |
| Max. P | 0.997347 |

| Location | 8,878,694 – 8,878,785 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 97.39 |
| Mean single sequence MFE | -23.36 |
| Consensus MFE | -20.94 |
| Energy contribution | -21.18 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.637673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
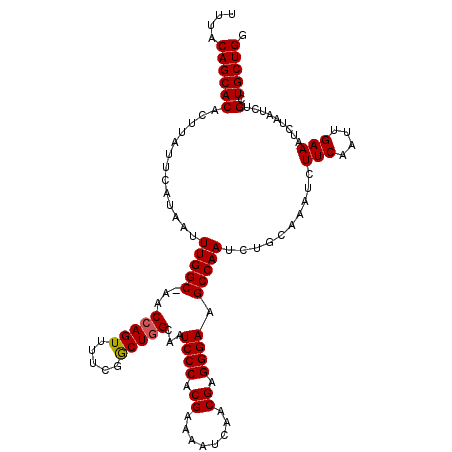
>X_DroMel_CAF1 8878694 91 + 22224390 AAAACCCUCCA--UUUCCCCAGCAGCAGAUUAGAUUUCAAUUGAAGAUUUGCAGAUUGGCUUCCCUCGUUGAUUUUCGUGGGAUUGCCAGCCG ...........--........((.(((((((....(((....))))))))))....((((.((((.((........)).))))..)))))).. ( -23.20) >DroSec_CAF1 9005 91 + 1 AAAACCCUCCA--UUUCCCCAGCAGCAGAUUAGAUUUCAAUUGAAGAUUUGCAGAUUGGCUUCCCUCGUUGAUUUUCGUGGGAUUGCCAGCCG ...........--........((.(((((((....(((....))))))))))....((((.((((.((........)).))))..)))))).. ( -23.20) >DroSim_CAF1 9321 91 + 1 AAAACCCUCCA--UUUCCCCAGCAGCAGAUUAGAUUUCAAUUGAAGAUUUGCAGAUUGGCUUCCCUCGUUGAUUUUCGUGGGAUUGCCAGCCG ...........--........((.(((((((....(((....))))))))))....((((.((((.((........)).))))..)))))).. ( -23.20) >DroEre_CAF1 8488 91 + 1 AAAACCCUCCA--UUUCCCCAGCAGCAGAUUAGAUUUCAAUUGAAGAUUUGCAGAUUGGCUUCCCUCGUUGAUUUUCGUGGGAUUGCCAGUCG ...........--...........(((((((....(((....)))))))))).(((((((.((((.((........)).))))..))))))). ( -27.30) >DroYak_CAF1 9411 93 + 1 AAAACCCUCCAUAUUUCCACAGCAGCCGAUUAGAUUUCAAUUGAAGAUUUGCAGAUUGGCUUCCCUCGUUGAUUUUCGUGGGAUUGCGAGCCG .....................(((((((((((((((((....).))))))...))))))).((((.((........)).)))).)))...... ( -19.90) >consensus AAAACCCUCCA__UUUCCCCAGCAGCAGAUUAGAUUUCAAUUGAAGAUUUGCAGAUUGGCUUCCCUCGUUGAUUUUCGUGGGAUUGCCAGCCG .....................((.(((((((....(((....))))))))))....((((.((((.((........)).))))..)))))).. (-20.94 = -21.18 + 0.24)


| Location | 8,878,710 – 8,878,824 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 97.88 |
| Mean single sequence MFE | -33.12 |
| Consensus MFE | -31.74 |
| Energy contribution | -31.98 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8878710 114 + 22224390 CCAGCAGCAGAUUAGAUUUCAAUUGAAGAUUUGCAGAUUGGCUUCCCUCGUUGAUUUUCGUGGGAUUGCCAGCCGAAAACUGGCUAGCCAAAUUAUGAAUAAGUGUGCUGUAAA .((((((((((((....(((....))))))))))...((((((((((.((........)).))))..(((((.......))))).))))))..............))))).... ( -35.90) >DroSec_CAF1 9021 113 + 1 CCAGCAGCAGAUUAGAUUUCAAUUGAAGAUUUGCAGAUUGGCUUCCCUCGUUGAUUUUCGUGGGAUUGCCAGCCGAAAACUGGUU-GCCAAAUUAUGAAUAAGUGUGCUGUAAA .((((((((((((....(((....))))))))))...(((((.((((.((........)).))))..(((((.......))))).-)))))..............))))).... ( -32.90) >DroSim_CAF1 9337 113 + 1 CCAGCAGCAGAUUAGAUUUCAAUUGAAGAUUUGCAGAUUGGCUUCCCUCGUUGAUUUUCGUGGGAUUGCCAGCCGAAAACUGGUU-GCCAAAUUAUGAAUAAGUGUGCUGUAAA .((((((((((((....(((....))))))))))...(((((.((((.((........)).))))..(((((.......))))).-)))))..............))))).... ( -32.90) >DroEre_CAF1 8504 113 + 1 CCAGCAGCAGAUUAGAUUUCAAUUGAAGAUUUGCAGAUUGGCUUCCCUCGUUGAUUUUCGUGGGAUUGCCAGUCGAAAACUGGUU-GCCAAAUUAUGAAUAAGUGUGCUGUAAA .((((((((((((....(((....))))))))))...(((((.((((.((........)).))))..((((((.....)))))).-)))))..............))))).... ( -34.50) >DroYak_CAF1 9429 113 + 1 ACAGCAGCCGAUUAGAUUUCAAUUGAAGAUUUGCAGAUUGGCUUCCCUCGUUGAUUUUCGUGGGAUUGCGAGCCGAAAACUGGUU-GCCAAAUUAUGAAUAAGUGUGCUGUAAA ((((((((((((((((((((....).))))))...))))))).((((.((........)).))))..((.(((((.....)))))-)).................))))))... ( -29.40) >consensus CCAGCAGCAGAUUAGAUUUCAAUUGAAGAUUUGCAGAUUGGCUUCCCUCGUUGAUUUUCGUGGGAUUGCCAGCCGAAAACUGGUU_GCCAAAUUAUGAAUAAGUGUGCUGUAAA .((((((((((((....(((....))))))))))...(((((.((((.((........)).))))..(((((.......)))))..)))))..............))))).... (-31.74 = -31.98 + 0.24)


| Location | 8,878,710 – 8,878,824 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 97.88 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -24.34 |
| Energy contribution | -24.38 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8878710 114 - 22224390 UUUACAGCACACUUAUUCAUAAUUUGGCUAGCCAGUUUUCGGCUGGCAAUCCCACGAAAAUCAACGAGGGAAGCCAAUCUGCAAAUCUUCAAUUGAAAUCUAAUCUGCUGCUGG ....(((((((............((((((.((((((.....))))))..((((.((........)).))))))))))..........(((....)))........)).))))). ( -32.80) >DroSec_CAF1 9021 113 - 1 UUUACAGCACACUUAUUCAUAAUUUGGC-AACCAGUUUUCGGCUGGCAAUCCCACGAAAAUCAACGAGGGAAGCCAAUCUGCAAAUCUUCAAUUGAAAUCUAAUCUGCUGCUGG ....(((((((............(((((-..(((((.....)))))...((((.((........)).)))).)))))..........(((....)))........)).))))). ( -27.70) >DroSim_CAF1 9337 113 - 1 UUUACAGCACACUUAUUCAUAAUUUGGC-AACCAGUUUUCGGCUGGCAAUCCCACGAAAAUCAACGAGGGAAGCCAAUCUGCAAAUCUUCAAUUGAAAUCUAAUCUGCUGCUGG ....(((((((............(((((-..(((((.....)))))...((((.((........)).)))).)))))..........(((....)))........)).))))). ( -27.70) >DroEre_CAF1 8504 113 - 1 UUUACAGCACACUUAUUCAUAAUUUGGC-AACCAGUUUUCGACUGGCAAUCCCACGAAAAUCAACGAGGGAAGCCAAUCUGCAAAUCUUCAAUUGAAAUCUAAUCUGCUGCUGG ....(((((((............(((((-..((((((...))))))...((((.((........)).)))).)))))..........(((....)))........)).))))). ( -27.50) >DroYak_CAF1 9429 113 - 1 UUUACAGCACACUUAUUCAUAAUUUGGC-AACCAGUUUUCGGCUCGCAAUCCCACGAAAAUCAACGAGGGAAGCCAAUCUGCAAAUCUUCAAUUGAAAUCUAAUCGGCUGCUGU ...((((((..(...........(((((-..((.......)).......((((.((........)).)))).)))))..........(((....)))........)..)))))) ( -21.80) >consensus UUUACAGCACACUUAUUCAUAAUUUGGC_AACCAGUUUUCGGCUGGCAAUCCCACGAAAAUCAACGAGGGAAGCCAAUCUGCAAAUCUUCAAUUGAAAUCUAAUCUGCUGCUGG ....((((((.............(((((...(((((.....)))))...((((.((........)).)))).)))))..........(((....))).........).))))). (-24.34 = -24.38 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:28 2006