
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,875,619 – 8,875,777 |
| Length | 158 |
| Max. P | 0.927056 |

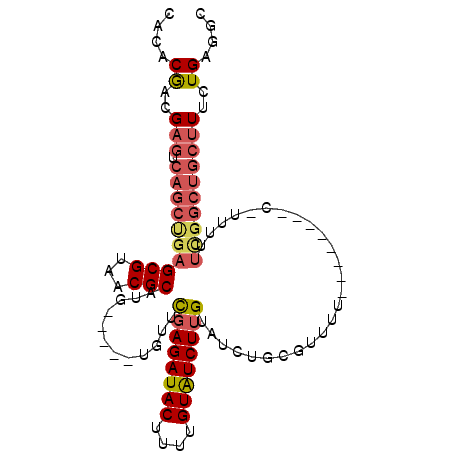
| Location | 8,875,619 – 8,875,718 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 81.33 |
| Mean single sequence MFE | -29.13 |
| Consensus MFE | -18.36 |
| Energy contribution | -19.50 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.876701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8875619 99 + 22224390 AUCUCUGCCUUUUGCUGGCGAGUGCUACAAUUGUCGCACGUAAUCGUGCUGGCAGUUACGCCUCAGAAAGCAGCCAAAAAAAAAAGAAAAAAAAACGCA ....((((.(((((..((((.(.....)((((((((((((....))))).))))))).)))).))))).)))).......................... ( -28.80) >DroSec_CAF1 6093 87 + 1 AUC--UGCCUUUUGCUGGCGAGUGCUACAAUUGUCGCACGUAAUCGUGCUGGCAGUUACGCCUCAGAAAGCAGCCGAAAA----------AAAAACGCA ..(--(((.(((((..((((.(.....)((((((((((((....))))).))))))).)))).))))).)))).......----------......... ( -28.80) >DroSim_CAF1 6442 88 + 1 AUC--UGCCUUUUGCUGGCGAGUGCUACAAUUGUCGCACGUAAUCGUGCUGGCAGUUACGCCUCAGAAAGCAGCCGAAAAA---------AAAAACGCA ..(--(((.(((((..((((.(.....)((((((((((((....))))).))))))).)))).))))).))))........---------......... ( -28.80) >DroEre_CAF1 6099 83 + 1 AUC--UGCCUUUC---AGCGAGUGCUACAAUUGUCGCACGUAAUCGUGCUGGCAGUUACGGCUCAGAAAGCACCGAAAAAGCG--------UAGAU--- (((--(((((((.---..((.(((((..((((((((((((....))))).))))))).(......)..)))))))..)))).)--------)))))--- ( -30.20) >DroYak_CAF1 6428 89 + 1 AUC--UGCCUUUUGCUGGCGAGUGCUACAAUUGUCGCACGUAAUCGUGCUGGCAGUUACGCCUCAGAAAGCAGCCGAAAAACG--------UAGAUACA (((--(((.((((..(((((((.((...((((((((((((....))))).)))))))..))))).(....).)))).)))).)--------)))))... ( -31.80) >DroAna_CAF1 10970 85 + 1 AUC--UGCCUG-CGGUGGCGAGUGUUACAGUUGUCGCACGUAAUAGCG----CAGUUACGCCUCAGAAAGCAGCCCGAAAAAGCU-------GAAGGCA ..(--((((((-(((((((((.((...)).))))))).)))...)).)----)))....((((.......((((........)))-------).)))). ( -26.40) >consensus AUC__UGCCUUUUGCUGGCGAGUGCUACAAUUGUCGCACGUAAUCGUGCUGGCAGUUACGCCUCAGAAAGCAGCCGAAAAA_G________AAAACGCA .....(((.((((...((((.(.....)((((((((((((....))))).))))))).))))...)))))))........................... (-18.36 = -19.50 + 1.14)

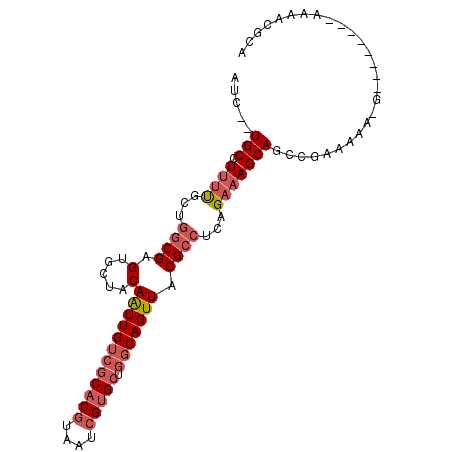

| Location | 8,875,678 – 8,875,777 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 78.12 |
| Mean single sequence MFE | -28.01 |
| Consensus MFE | -16.22 |
| Energy contribution | -17.08 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8875678 99 - 22224390 CACACGACGAGUCAGCUGAGCGUAACGCAUG-----UGUUCGAGAUACUUUUGUAUCUUGUAUCUGCGUUUUUUUUUCUUUUUUUUUUGGCUGCUUUCUGAGGC ..((.((.(((.((((..((...((((((.(-----((..((((((((....))))))))))).))))))...............))..))))))))))).... ( -25.67) >DroSec_CAF1 6150 89 - 1 CACACGACGAGUCAGCUGAGCGUAACGCAUG-----UGUUCGAGAUACUUUUGUAUCUUGUAUCUGCGUUUUU----------UUUUCGGCUGCUUUCUGAGGC ..((.((.(((.((((((((...((((((.(-----((..((((((((....))))))))))).))))))...----------..))))))))))))))).... ( -30.90) >DroSim_CAF1 6499 90 - 1 CACACGACGAGUCAGCUGAGCGUAACGCAUG-----UGUUCGAGAUACUUUUGUAUCUUGUAUCUGCGUUUUU---------UUUUUCGGCUGCUUUCUGAGGC ..((.((.(((.((((((((...((((((.(-----((..((((((((....))))))))))).))))))...---------...))))))))))))))).... ( -30.30) >DroEre_CAF1 6153 85 - 1 CACACGGCGAGUCAGCUGAGCGUAACGCAUG-----UGUUUGAGAUACUUUUGUAUCUUGU------AUCUA--------CGCUUUUUCGGUGCUUUCUGAGCC .....(((.((..(((.((((((((((....-----)))..(((((((....)))))))..------...))--------))))).......)))..))..))) ( -22.40) >DroYak_CAF1 6485 91 - 1 CACACGGCGAGUCAGCUGAGCGUAACGCAUG-----UGUUUGAGAUACUUUUGUGUCUUGUAUCUGUAUCUA--------CGUUUUUCGGCUGCUUUCUGAGGC ....(((.(((.((((((((((((..(((.(-----((..((((((((....))))))))))).)))...))--------)))...)))))))))).))).... ( -29.00) >DroAna_CAF1 11022 89 - 1 CACCCAACG-----GACGAGCGUCACGCAUGAAGAGUGCCUCAGAUACUUUUGUAUCUCGG---UGCCUUC-------AGCUUUUUCGGGCUGCUUUCUGAGGC ..(((....-----(((....)))..((.(((((.(..((..((((((....)))))).))---..)))))-------)))......)))..((((....)))) ( -29.80) >consensus CACACGACGAGUCAGCUGAGCGUAACGCAUG_____UGUUCGAGAUACUUUUGUAUCUUGUAUCUGCGUUUU________C_UUUUUCGGCUGCUUUCUGAGGC ....((..(((.((((((((((...)))............((((((((....))))))))..........................))))))))))..)).... (-16.22 = -17.08 + 0.86)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:20 2006