| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,051,262 – 1,051,374 |
| Length | 112 |
| Max. P | 0.548682 |

| Location | 1,051,262 – 1,051,374 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.08 |
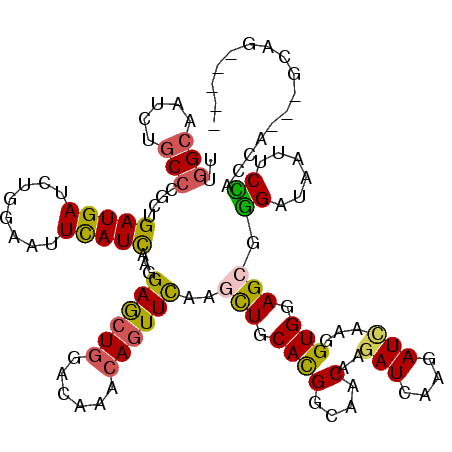
| Mean single sequence MFE | -33.17 |
| Consensus MFE | -20.08 |
| Energy contribution | -19.67 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1051262 112 + 22224390 UUGGCAAUCUGCCGGCGGAUGACCUGGAAUUCAUUAAGGAGCUGGACAAGCAGUUCAAACUGCACGGAAACAAGAUCAAGAUCAAGGUGGAGAGGGAUAAUUCCACGA---GUAG----- ..((..(((((....)))))..))(((((((.(((...((((((......))))))...((.((((....)..(((....)))...))).))...))))))))))...---....----- ( -30.50) >DroVir_CAF1 302 117 + 1 UUGGCAAUUUGCCCGCCGAUGAUCUGGAGUUCAUCAAGGAGCUAGACAAACAGUUUAAGCUACACGGCAACAAGAUCAAAAUUAAGGUGGAGCGUGAGAAUACGACCA---GCAGUAACG ..(((.....))).((((((((........)))))..(.((((((((.....)))).)))).)..))).................(.(((..((((....)))).)))---.)....... ( -27.70) >DroGri_CAF1 260 117 + 1 UUGGCAAUUUGCCCGCUGAUGAUCUGGAAUUCAUCAAAGAGCUGGACAAACAGUUCAAGCUGCACGGCAGCAAGAUCAAAAUCAAAGUGGAGCGUGAUAAUUCAACCA---GCAGCAACG ..(((.....))).((((.((((((.............((((((......))))))..(((((...))))).)))))).........(((....(((....))).)))---.)))).... ( -35.40) >DroMoj_CAF1 264 120 + 1 UUGGCAAUUUGCCCGCUGAUGAUCUGGAGUUCAUCAAGGAGCUGGACAAACAGUUCAAGCUGCACGGCAACAAGAUCAAGAUCAAAGUGGAGCGUGAGAAUACGACCACCAGCAGCAGCA ..(((.....))).((((((((........)))))...((((((......))))))..(((((..(....)..(((....)))...((((..((((....)))).))))..)))))))). ( -39.20) >DroAna_CAF1 267 112 + 1 UCGGGAAUCUGCCAGCAGAUGACUUGGAAUUUAUUAAGGAACUGGACAAGCAAUUCAAGUUGCACGGAAACAAAAUCAAGAUCAAGGUGGAGAGGGAUAAUUCCACAU---CCAG----- (((((.(((((....)))))..)))))..............(((((...(((((....)))))..(....)...............((((((........)))))).)---))))----- ( -27.20) >DroPer_CAF1 5143 112 + 1 UUAGCAACCUGCCCGCUGAUGAUCUGGAGUUCAUCAAGGAGCUGGACAAGCAGUUCAAGCUGCAUGGGGACAAGAUCAAGAUCAAGGUGGAGCGGGACAAUUCCACGG---GUAG----- .......((((((((((..((((((.((((((.(((.(.((((((((.....)))).)))).).)))))))....)).)))))).))))).)))))............---....----- ( -39.00) >consensus UUGGCAAUCUGCCCGCUGAUGAUCUGGAAUUCAUCAAGGAGCUGGACAAACAGUUCAAGCUGCACGGCAACAAGAUCAAGAUCAAGGUGGAGCGGGAUAAUUCCACCA___GCAG_____ ..(((.....)))....(((((........)))))...((((((......))))))..(((.((((....)..(((....)))...))).))).((......))................ (-20.08 = -19.67 + -0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:56 2006