
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,865,379 – 8,865,494 |
| Length | 115 |
| Max. P | 0.991069 |

| Location | 8,865,379 – 8,865,494 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -17.88 |
| Energy contribution | -18.68 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
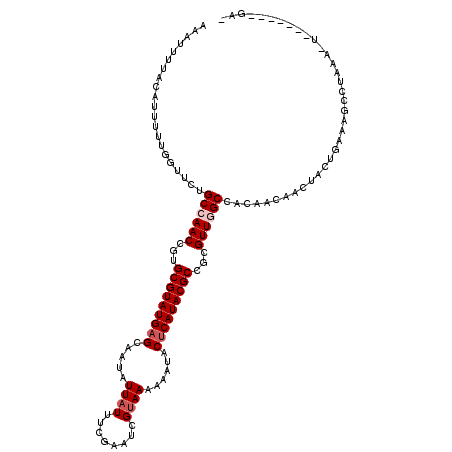
>X_DroMel_CAF1 8865379 115 + 22224390 AAAUUUUACAUU-UUGGUUUUGCCAACCGUGCGUAUGAGCAAUAUUAUUGCGAAUCGUAAAAAUUACUCAUACGCCGCGUUAGCCACAUAAUCUACUGAAAGCCUAAACUAU----CGAG ............-..((((((((.(((((.(((((((((....(((.(((((...))))).)))..))))))))))).))).))..((........))))))))........----.... ( -26.80) >DroSec_CAF1 772 106 + 1 AAAUUUUAAAUUUUUGGUUCUGCCAACUGUGCGUAUGAGCAAUAUUAUUUCGAAUCGUAAAAAAUACUCAUACGCCGCGUUGGCCACAACAACUCGUGAAUGCCUA-------------- ...............(((...((((((((.(((((((((...((((.(((((...)).))).))))))))))))))).))))))(((........)))...)))..-------------- ( -27.70) >DroSim_CAF1 571 103 + 1 AAAUUUUACAUUUUUGGUUCUGCCAACCGUGCGUAUGAGCAAUAUUAUUUCGAAUCGUAAAAAAUACUCAUACGCCGCGUUGGCCAC---AACUCGUGAAAGCCUA-------------- ...............((((..((((((((.(((((((((...((((.(((((...)).))).))))))))))))))).))))))(((---.....)))..))))..-------------- ( -30.60) >DroEre_CAF1 898 117 + 1 AAAUUUG--AUUUUUGGUUCUGCCAACCGUGCGUAUGAGCAAUAUUAUUACGAAUCGUAA-AAAUACUCAUACGCCCUGUUAGCCACAACAACUACUGAAAGCUUAAAGUCAAUGCAGAC ....(((--(((((.((((..((.(((.(.(((((((((...((((.(((((...)))))-.))))))))))))).).))).)).....((.....))..)))).))))))))....... ( -28.20) >DroYak_CAF1 844 115 + 1 AAAUUUA-CAAUUUCGGUUCUGCCAACCAUGCGUAUGAGCAAUAUUAUUUCGAAUUGCAAAAAAUACGCAUACGCCUCGUUGGCCACAACAACUACUGAAAGCUUCAAUUCU----UGAG .......-...(((((((...((((((...(((((((.(((((..........)))))..........)))))))...))))))..........)))))))...........----.... ( -24.12) >consensus AAAUUUUACAUUUUUGGUUCUGCCAACCGUGCGUAUGAGCAAUAUUAUUUCGAAUCGUAAAAAAUACUCAUACGCCGCGUUGGCCACAACAACUACUGAAAGCCUAAA_U_______GA_ .....................((((((...(((((((((.....((((........))))......)))))))))...)))))).................................... (-17.88 = -18.68 + 0.80)

| Location | 8,865,379 – 8,865,494 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -25.50 |
| Energy contribution | -25.46 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8865379 115 - 22224390 CUCG----AUAGUUUAGGCUUUCAGUAGAUUAUGUGGCUAACGCGGCGUAUGAGUAAUUUUUACGAUUCGCAAUAAUAUUGCUCAUACGCACGGUUGGCAAAACCAA-AAUGUAAAAUUU ...(----(.(((....))).))..((.(((..((.((((((.(((((((((((((((...................))))))))))))).))))))))...))...-))).))...... ( -31.81) >DroSec_CAF1 772 106 - 1 --------------UAGGCAUUCACGAGUUGUUGUGGCCAACGCGGCGUAUGAGUAUUUUUUACGAUUCGAAAUAAUAUUGCUCAUACGCACAGUUGGCAGAACCAAAAAUUUAAAAUUU --------------..((....(((((....)))))((((((...(((((((((((.......................)))))))))))...))))))....))............... ( -32.50) >DroSim_CAF1 571 103 - 1 --------------UAGGCUUUCACGAGUU---GUGGCCAACGCGGCGUAUGAGUAUUUUUUACGAUUCGAAAUAAUAUUGCUCAUACGCACGGUUGGCAGAACCAAAAAUGUAAAAUUU --------------..((....(((.....---)))((((((.(((((((((((((.......................))))))))))).))))))))....))............... ( -31.90) >DroEre_CAF1 898 117 - 1 GUCUGCAUUGACUUUAAGCUUUCAGUAGUUGUUGUGGCUAACAGGGCGUAUGAGUAUUU-UUACGAUUCGUAAUAAUAUUGCUCAUACGCACGGUUGGCAGAACCAAAAAU--CAAAUUU (((......)))....((((......))))(((.((.(((((...(((((((((((...-(((((...)))))......)))))))))))...))))))).))).......--....... ( -33.60) >DroYak_CAF1 844 115 - 1 CUCA----AGAAUUGAAGCUUUCAGUAGUUGUUGUGGCCAACGAGGCGUAUGCGUAUUUUUUGCAAUUCGAAAUAAUAUUGCUCAUACGCAUGGUUGGCAGAACCGAAAUUG-UAAAUUU (.((----(.(((((..(....)..))))).))).)((((((.(.(((((((.(((..(((((.....)))))......))).))))))).).)))))).............-....... ( -27.60) >consensus _UC_______A_UUUAGGCUUUCAGUAGUUGUUGUGGCCAACGCGGCGUAUGAGUAUUUUUUACGAUUCGAAAUAAUAUUGCUCAUACGCACGGUUGGCAGAACCAAAAAUGUAAAAUUU .................................((.((((((...(((((((((((.......................)))))))))))...))))))...))................ (-25.50 = -25.46 + -0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:16 2006