
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,839,543 – 8,839,645 |
| Length | 102 |
| Max. P | 0.688559 |

| Location | 8,839,543 – 8,839,645 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.32 |
| Mean single sequence MFE | -31.97 |
| Consensus MFE | -21.67 |
| Energy contribution | -23.15 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
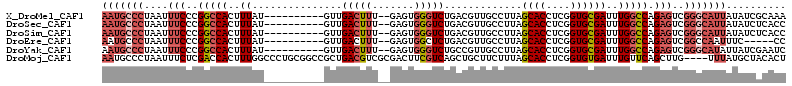
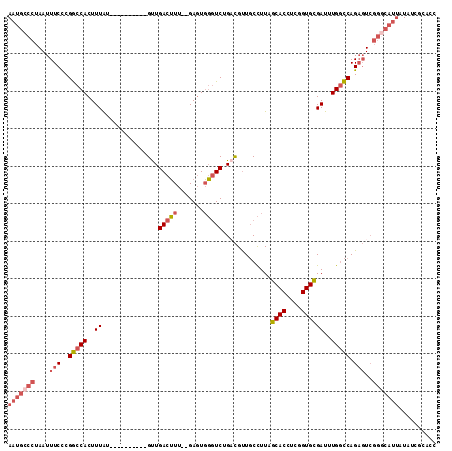
>X_DroMel_CAF1 8839543 102 + 22224390 AAUGCCCUAAUUUCCCGGCCACUUUAU----------GUUGACUUU--GAGUGGGUCUGACGUUGCCUUAGCACCUCGGUGCGAUUUGGCCAGAGUCGGGCAUUAUAUCGCAAA (((((((.........((((((((...----------(....)...--)))).)))).(((...(((...((((....)))).....)))....)))))))))).......... ( -33.80) >DroSec_CAF1 79814 102 + 1 AAUGCCCUAAUUUCCCGGCCACUUUAU----------GUUGACUUU--GAGUGGGUCUGACGUUGCCUUAGCACCUCGGUGCGAUUUGGCCAGAGUCGGGCAUUAUAUCUCACC (((((((.........((((((((...----------(....)...--)))).)))).(((...(((...((((....)))).....)))....)))))))))).......... ( -33.80) >DroSim_CAF1 71613 102 + 1 AAUGCCCUAAUUUCCCGGCCACUUUAU----------GUUGACUUU--GAGUGGGUCUGACGUUGCCUUAGCACCUCGGUGCGAUUUGGCCAGAGUCGGGCAUUAUAUCUCACC (((((((.........((((((((...----------(....)...--)))).)))).(((...(((...((((....)))).....)))....)))))))))).......... ( -33.80) >DroEre_CAF1 74982 97 + 1 AAUGCCCUAAUUUCCCGGCCACUUUAU----------GUUGACUUU--GAGUGGCUCUGACGUUGCCUUAGCACCUCGGUGCGAUUUGGCCAGAGUCGGCCAAUUUC-----CC ................((((((((...----------(....)...--))))(((((((.....(((...((((....)))).....))))))))))))))......-----.. ( -31.10) >DroYak_CAF1 76581 102 + 1 AAUGCCCUAAUUUCCCGGCCACUUUAU----------GUUGACUUU--GAGUGGGUCUGCCGUUGCCUUAGCACCUCGGUGCGAUUUGGCCAGAGUCGGGCAUAUUAUCGAAUC .((((((....(((..(((((..((..----------((.(((((.--....))))).))..........((((....))))))..))))).)))..))))))........... ( -31.40) >DroMoj_CAF1 121283 110 + 1 AAUGCCCUAAUUUCUCGACCACUUUGGCCCUGCGGCCGCUGACGUCGCGACUUCGUCAGCUGCUUCUUUAGCACCUCGGUGUGAUUUGUUCAGCUUG----UUUAUGCUACACU ...............(((.......((((....))))(((((((.........)))))))((((.....))))..)))((((((.....))(((...----.....))))))). ( -27.90) >consensus AAUGCCCUAAUUUCCCGGCCACUUUAU__________GUUGACUUU__GAGUGGGUCUGACGUUGCCUUAGCACCUCGGUGCGAUUUGGCCAGAGUCGGGCAUUAUAUCGCACC (((((((....(((..(((((..((...............(((((.......))))).............((((....))))))..))))).)))..))))))).......... (-21.67 = -23.15 + 1.48)

| Location | 8,839,543 – 8,839,645 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 81.32 |
| Mean single sequence MFE | -30.96 |
| Consensus MFE | -21.46 |
| Energy contribution | -23.35 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
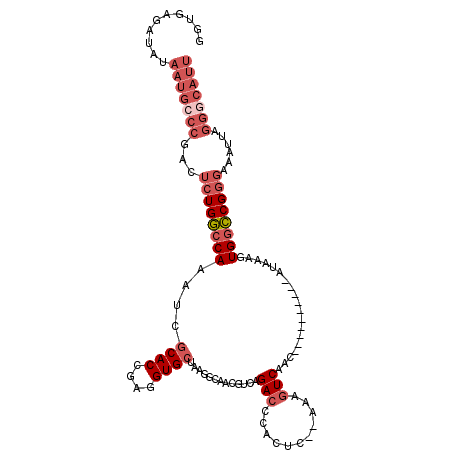
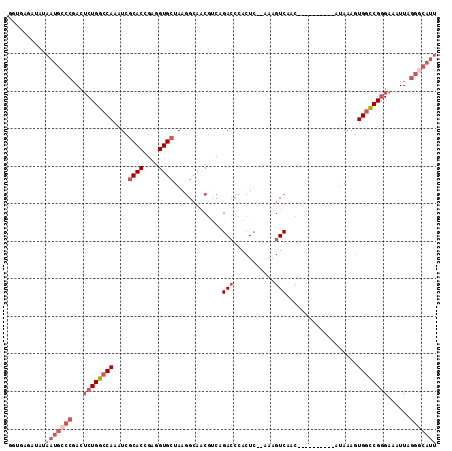
>X_DroMel_CAF1 8839543 102 - 22224390 UUUGCGAUAUAAUGCCCGACUCUGGCCAAAUCGCACCGAGGUGCUAAGGCAACGUCAGACCCACUC--AAAGUCAAC----------AUAAAGUGGCCGGGAAAUUAGGGCAUU ..........(((((((...((((((((....((((....))))...(....)....(((......--...)))...----------......))))))))......))))))) ( -31.70) >DroSec_CAF1 79814 102 - 1 GGUGAGAUAUAAUGCCCGACUCUGGCCAAAUCGCACCGAGGUGCUAAGGCAACGUCAGACCCACUC--AAAGUCAAC----------AUAAAGUGGCCGGGAAAUUAGGGCAUU ..........(((((((...((((((((....((((....))))...(....)....(((......--...)))...----------......))))))))......))))))) ( -31.70) >DroSim_CAF1 71613 102 - 1 GGUGAGAUAUAAUGCCCGACUCUGGCCAAAUCGCACCGAGGUGCUAAGGCAACGUCAGACCCACUC--AAAGUCAAC----------AUAAAGUGGCCGGGAAAUUAGGGCAUU ..........(((((((...((((((((....((((....))))...(....)....(((......--...)))...----------......))))))))......))))))) ( -31.70) >DroEre_CAF1 74982 97 - 1 GG-----GAAAUUGGCCGACUCUGGCCAAAUCGCACCGAGGUGCUAAGGCAACGUCAGAGCCACUC--AAAGUCAAC----------AUAAAGUGGCCGGGAAAUUAGGGCAUU ..-----....(((((((....)))))))...((.((((.((((....)).)).)).(.((((((.--.........----------....))))))).........))))... ( -28.74) >DroYak_CAF1 76581 102 - 1 GAUUCGAUAAUAUGCCCGACUCUGGCCAAAUCGCACCGAGGUGCUAAGGCAACGGCAGACCCACUC--AAAGUCAAC----------AUAAAGUGGCCGGGAAAUUAGGGCAUU ...........((((((...((((((((....((((....))))...(....)....(((......--...)))...----------......))))))))......)))))). ( -30.90) >DroMoj_CAF1 121283 110 - 1 AGUGUAGCAUAAA----CAAGCUGAACAAAUCACACCGAGGUGCUAAAGAAGCAGCUGACGAAGUCGCGACGUCAGCGGCCGCAGGGCCAAAGUGGUCGAGAAAUUAGGGCAUU .((.((((.....----...)))).))..(((((.......((((.....))))(((((((.........)))))))((((....))))...)))))................. ( -31.00) >consensus GGUGAGAUAUAAUGCCCGACUCUGGCCAAAUCGCACCGAGGUGCUAAGGCAACGUCAGACCCACUC__AAAGUCAAC__________AUAAAGUGGCCGGGAAAUUAGGGCAUU ..........(((((((...((((((((....((((....)))).............(((...........)))...................))))))))......))))))) (-21.46 = -23.35 + 1.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:05 2006