
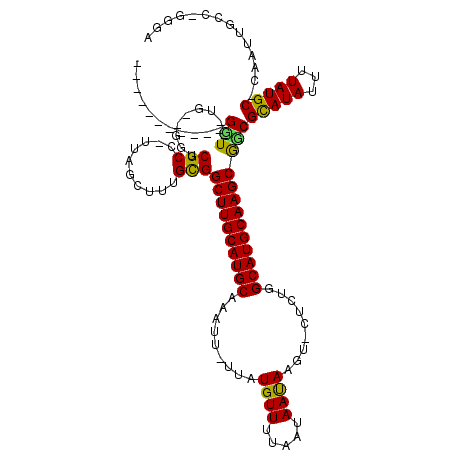
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,827,461 – 8,827,561 |
| Length | 100 |
| Max. P | 0.955332 |

| Location | 8,827,461 – 8,827,561 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.87 |
| Mean single sequence MFE | -33.39 |
| Consensus MFE | -22.61 |
| Energy contribution | -21.67 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8827461 100 + 22224390 -----------GGU--UA--GGUCGCC-AUGGCUUUGCGGCUUGCAUGCAAAUU-UUAUGUUUUAAUAAUAAGU-CUCUGGCAUGCAAGCGCCGCAUAUUUUAUGC-CAAUUGAU-GGAG -----------(((--..--....)))-.((((..((((((((((((((....(-((((.........))))).-.....))))))))..))))))........))-))......-.... ( -29.70) >DroVir_CAF1 106081 111 + 1 CUUUGGCGUCUGCU--UG--AGUCGC--UUAGCUUUGCGGCUUGCAUGCAAAUU-UUAUGUUUUAAUAACAAGUUCUCUGGCAUGCAAGCGGCGCAUAUUUUAUGC-CAAUUGCC-GUGC ....((((.(((((--((--((((((--........)))))))((((((.....-...((((.....)))).........)))))))))))))(((((...)))))-.....)))-.... ( -36.73) >DroWil_CAF1 13655 105 + 1 -----------GGU--UGUUGUGCGCCUUUAGGUUUGCGGCUUGCAUGCAAAUUUUUAUGUUUUAAUAAUAAGU-CUGAGGCAUGCAAGCAGCGUAUAUUUUAUGC-CAAUUGCCUGGUA -----------(((--.((((.(((((....)))..((.((((((((((.....(((((.........))))).-.....)))))))))).))...........))-)))).)))..... ( -33.70) >DroMoj_CAF1 109110 110 + 1 UUUUGGCGUCUGCU--UG--AGUCGC--UUAGCUUUGUGGCUUGCAUGCAAAUU-UUAUGUUUUAAUAACAAGU-CUCUGGCAUGCAAGCGGCGCAUAUUUUAUGC-CAAUUGCC-GUGC ....((((.(((((--((--((((((--........)))))))((((((.....-...((((.....))))...-.....)))))))))))))(((((...)))))-.....)))-.... ( -35.39) >DroAna_CAF1 56662 98 + 1 -----------GGU--UG--GGUCGCC---GGCUCUGCGGCUUGCAUGCAAAUU-UUAUGUUUUAAUAAUAAGU-CUCUGGCAUGCAAGCGUCGCAUAUUUUAUGC-CGAUUGCU-GCUG -----------(((--..--....)))---(((...(((((((((((((....(-((((.........))))).-.....))))))))))((((((((...)))).-))))))).-))). ( -30.40) >DroPer_CAF1 69762 104 + 1 -----------GGUUUUG--GGUCGCCUCUGGCUUUGCGGCUUGCAUGCAAAUU-UUAUGUUUUAAUAAUAAGU-CUGUGGCAUGCAAGCAUCGCAUAUUUUAUGCCCAAUUGCU-GGGA -----------(((.(((--(((.(((...)))..((((((((((((((....(-((((.........))))).-.....))))))))))..))))........))))))..)))-.... ( -34.40) >consensus ___________GGU__UG__GGUCGCC_UUAGCUUUGCGGCUUGCAUGCAAAUU_UUAUGUUUUAAUAAUAAGU_CUCUGGCAUGCAAGCGGCGCAUAUUUUAUGC_CAAUUGCC_GGGA ...........(((.........(((..........)))((((((((((.........((((.....)))).........)))))))))))))(((((...))))).............. (-22.61 = -21.67 + -0.94)


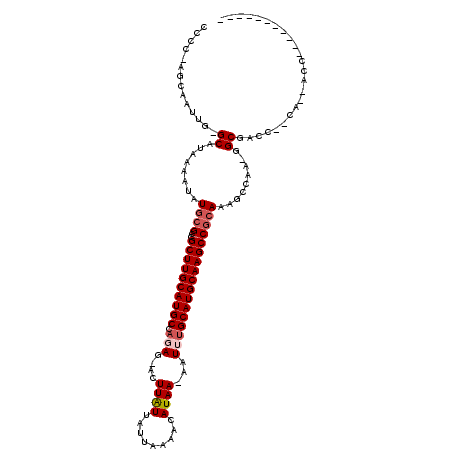
| Location | 8,827,461 – 8,827,561 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.87 |
| Mean single sequence MFE | -28.46 |
| Consensus MFE | -18.75 |
| Energy contribution | -19.36 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8827461 100 - 22224390 CUCC-AUCAAUUG-GCAUAAAAUAUGCGGCGCUUGCAUGCCAGAG-ACUUAUUAUUAAAACAUAA-AAUUUGCAUGCAAGCCGCAAAGCCAU-GGCGACC--UA--ACC----------- (.((-((......-(((((...)))))(((((((((((((.(((.-..((((.........))))-..)))))))))))))......)))))-)).)...--..--...----------- ( -25.80) >DroVir_CAF1 106081 111 - 1 GCAC-GGCAAUUG-GCAUAAAAUAUGCGCCGCUUGCAUGCCAGAGAACUUGUUAUUAAAACAUAA-AAUUUGCAUGCAAGCCGCAAAGCUAA--GCGACU--CA--AGCAGACGCCAAAG ....-(((.....-(((((...)))))((.((((((((((.(((.....((((.....))))...-..))))))))))))).))........--((....--..--.))....))).... ( -34.50) >DroWil_CAF1 13655 105 - 1 UACCAGGCAAUUG-GCAUAAAAUAUACGCUGCUUGCAUGCCUCAG-ACUUAUUAUUAAAACAUAAAAAUUUGCAUGCAAGCCGCAAACCUAAAGGCGCACAACA--ACC----------- .....((...(((-((...........((.((((((((((.....-.........................)))))))))).))...((....)).)).)))..--.))----------- ( -23.11) >DroMoj_CAF1 109110 110 - 1 GCAC-GGCAAUUG-GCAUAAAAUAUGCGCCGCUUGCAUGCCAGAG-ACUUGUUAUUAAAACAUAA-AAUUUGCAUGCAAGCCACAAAGCUAA--GCGACU--CA--AGCAGACGCCAAAA ....-(((..(((-(((((...)))))...((((((((((.(((.-...((((.....))))...-..)))))))))))))..)))......--((....--..--.))....))).... ( -31.40) >DroAna_CAF1 56662 98 - 1 CAGC-AGCAAUCG-GCAUAAAAUAUGCGACGCUUGCAUGCCAGAG-ACUUAUUAUUAAAACAUAA-AAUUUGCAUGCAAGCCGCAGAGCC---GGCGACC--CA--ACC----------- ....-.((...((-((........((((..((((((((((.(((.-..((((.........))))-..)))))))))))))))))..)))---)))....--..--...----------- ( -26.40) >DroPer_CAF1 69762 104 - 1 UCCC-AGCAAUUGGGCAUAAAAUAUGCGAUGCUUGCAUGCCACAG-ACUUAUUAUUAAAACAUAA-AAUUUGCAUGCAAGCCGCAAAGCCAGAGGCGACC--CAAAACC----------- ....-.....((((((........((((..((((((((((.....-...................-.....))))))))))))))..(((...)))).))--)))....----------- ( -29.55) >consensus CCCC_AGCAAUUG_GCAUAAAAUAUGCGCCGCUUGCAUGCCAGAG_ACUUAUUAUUAAAACAUAA_AAUUUGCAUGCAAGCCGCAAAGCCAA_GGCGACC__CA__ACC___________ ..............((........((((..((((((((((.(((....((((.........))))...))))))))))))))))).........))........................ (-18.75 = -19.36 + 0.61)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:02 2006