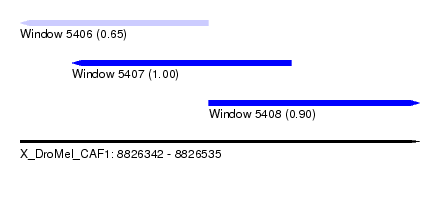
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,826,342 – 8,826,535 |
| Length | 193 |
| Max. P | 0.997921 |

| Location | 8,826,342 – 8,826,433 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 81.35 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -11.77 |
| Energy contribution | -13.03 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8826342 91 - 22224390 GAAAACUGGCACAGGAAAC-----------UGGCUAUUAAAAAGCAUUCUGCGAAAGUAGAUGUCAGGGGUCAGAGAAAAGUUGACUGCAGGGCUCAAAAAG .....(((((((((....)-----------))(((.......)))..(((((....)))))))))))(((((...(...((....)).)..)))))...... ( -28.20) >DroSec_CAF1 67605 91 - 1 GGCAACUGGCACAGGAAAC-----------UGGCUAUUAAAAAGCAUCCAGCGAAAAUAGAUGUCAGGGGUCCGAGAAAAGUUGACUGCAGGGCUCAAAAAG .....(((((((((....)-----------)).(((((.....((.....))...))))).))))))((((((..(...((....)).).))))))...... ( -25.50) >DroSim_CAF1 59659 91 - 1 GGCAACUGGCACAGGAAAC-----------UGGCUAUUAAAAAGCAUCCAGCGAAAAUAGAUGUCAGGGGUCCGAGAAAAGUUGACUGCAGGGCUCAAAAAG .....(((((((((....)-----------)).(((((.....((.....))...))))).))))))((((((..(...((....)).).))))))...... ( -25.50) >DroEre_CAF1 63149 93 - 1 GGAAAAUGGAAAUGGAAACAGGUAGGCGAAUGGCUUUUA-AAAGCAUCCUGCGAAAAG-AAUGUCUGGGGUCAGAGAAAAGUUGACUGCAGGACU------- ............((....)).(((((......((((...-.))))..)))))......-....((((.((((((.......)))))).))))...------- ( -22.00) >consensus GGAAACUGGCACAGGAAAC___________UGGCUAUUAAAAAGCAUCCAGCGAAAAUAGAUGUCAGGGGUCAGAGAAAAGUUGACUGCAGGGCUCAAAAAG .....((((((..(....)..............(((((.....((.....))...))))).))))))((((((......((....))...))))))...... (-11.77 = -13.03 + 1.25)



| Location | 8,826,367 – 8,826,473 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.10 |
| Mean single sequence MFE | -30.94 |
| Consensus MFE | -12.21 |
| Energy contribution | -13.74 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.39 |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.997921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8826367 106 - 22224390 UAGUCAUUUCAGAGGAUGCCAUGGCAACCACGCCUAAAAUGAAAACUGGCACAGGAAAC-----------UGGCUAUUAAAAAGCAUUCUGCGAAAGUAGAUGUCAGGGGUCAGAGA ......((((...((.(((....))).))..........(((...(((((((((....)-----------))(((.......)))..(((((....)))))))))))...))))))) ( -34.00) >DroSec_CAF1 67630 106 - 1 UAGGCACUGCAGAGGAUGCCAUGGCAACCACGCCUAAAAUGGCAACUGGCACAGGAAAC-----------UGGCUAUUAAAAAGCAUCCAGCGAAAAUAGAUGUCAGGGGUCCGAGA (((((.(....).((.(((....))).))..)))))...((((..(((((((((....)-----------)).(((((.....((.....))...))))).))))))..).)))... ( -33.80) >DroSim_CAF1 59684 106 - 1 UAGUCACUGCAGAGGAUGCCAUGGCAACCACGCCUAAAAUGGCAACUGGCACAGGAAAC-----------UGGCUAUUAAAAAGCAUCCAGCGAAAAUAGAUGUCAGGGGUCCGAGA .............((((.((.(((((.....(((......)))..((((..(((....)-----------))(((.......)))..))))..........))))))).)))).... ( -33.00) >DroEre_CAF1 63167 104 - 1 -----------GCGGAUGCCAUGGCAACCAAGUCCAAAAUGGAAAAUGGAAAUGGAAACAGGUAGGCGAAUGGCUUUUA-AAAGCAUCCUGCGAAAAG-AAUGUCUGGGGUCAGAGA -----------...(((.(((.((((.(((..(((.....)))...)))...((....)).(((((......((((...-.))))..)))))......-..))))))).)))..... ( -29.20) >DroYak_CAF1 63910 88 - 1 UAGUCAUUUCAGCGGAUGCCAUGGCAACCACGGCCAAA-U---------------------------GGAUGGCUAUUAAAAAGCAUCCUGCGAAAAG-GAUGUCAGAGGUCCAAGC .....(((((.(.((.(((....))).)).)(((((..-.---------------------------...)))))........(((((((......))-)))))..)))))...... ( -24.70) >consensus UAGUCACUGCAGAGGAUGCCAUGGCAACCACGCCUAAAAUGGAAACUGGCACAGGAAAC___________UGGCUAUUAAAAAGCAUCCUGCGAAAAUAGAUGUCAGGGGUCCGAGA .............((((.((.(((((.(((.(((.............)))...(....)...........)))..........((.....)).........))))))).)))).... (-12.21 = -13.74 + 1.53)

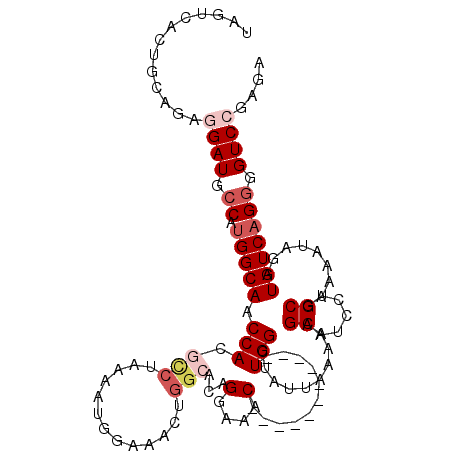

| Location | 8,826,433 – 8,826,535 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.11 |
| Mean single sequence MFE | -29.16 |
| Consensus MFE | -21.45 |
| Energy contribution | -21.21 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8826433 102 + 22224390 AUUUUAGGCGUGGUUGCCAUGGCAUCCUCUGAAAUGACUAGAUGCAUGUGCAGCCUGAAAAGAGUUCGAAUCGUAAUUAAGUCGCCAGUACA------------------ACGAAUUCGA .((((((((.....(((((((.((((.((......))...)))))))).))))))))))).(((((((....(((.............))).------------------.))))))).. ( -26.12) >DroSec_CAF1 67696 102 + 1 AUUUUAGGCGUGGUUGCCAUGGCAUCCUCUGCAGUGCCUAGGUGCAUUUGCAGCCUGAAAAGAGUUCGAAUCGUAAUUAAGUCGCCAUUACA------------------ACGAAUUCGA .(((((((((.((.(((....))).)).)((((((((......))).))))))))))))).(((((((....(((((.........))))).------------------.))))))).. ( -33.00) >DroSim_CAF1 59750 102 + 1 AUUUUAGGCGUGGUUGCCAUGGCAUCCUCUGCAGUGACUAGGUGCAUUUGCAGCCUGAAAAGAGUUCGAAUCGUAAUUAAGUCGCCAUUACA------------------ACGAAUUCGA .(((((((((.((.(((....))).)).)(((((((.(.....))).))))))))))))).(((((((....(((((.........))))).------------------.))))))).. ( -30.20) >DroEre_CAF1 63242 79 + 1 AUUUUGGACUUGGUUGCCAUGGCAUCCGC-------------UGCAUGUGCAGUCUGAAAAGAGUUCGA----------AGUCGUCAAUGCA------------------ACGAAUUUGA .((((((((((((.(((....))).))((-------------(((....))))).......))))))))----------))((((.......------------------))))...... ( -23.70) >DroYak_CAF1 63959 119 + 1 A-UUUGGCCGUGGUUGCCAUGGCAUCCGCUGAAAUGACUAGCUGCAUGUGCAGCCUGAAAAGAGUUCGAACCAUAAUUAAGUCGACAGUACAACGAAAUCAACGAUUUCAACGAAUUCAA (-(((((((((((...)))))))....((((...(((((.(((((....)))))((....)).................))))).)))).....((((((...))))))..))))).... ( -32.80) >consensus AUUUUAGGCGUGGUUGCCAUGGCAUCCUCUGAAAUGACUAGGUGCAUGUGCAGCCUGAAAAGAGUUCGAAUCGUAAUUAAGUCGCCAGUACA__________________ACGAAUUCGA .((((((((..((.(((....))).))...............(((....))))))))))).(((((((...........................................))))))).. (-21.45 = -21.21 + -0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:00 2006