
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,778,359 – 8,778,463 |
| Length | 104 |
| Max. P | 0.998840 |

| Location | 8,778,359 – 8,778,463 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.76 |
| Mean single sequence MFE | -29.77 |
| Consensus MFE | -18.08 |
| Energy contribution | -18.63 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
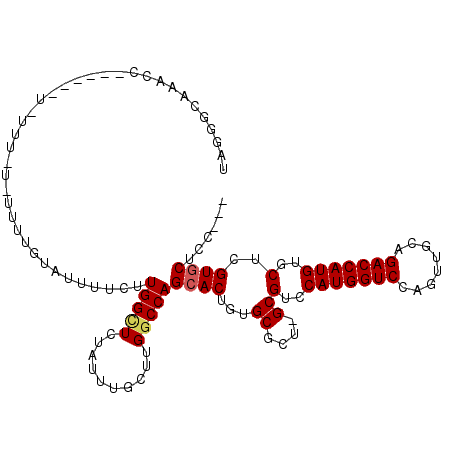
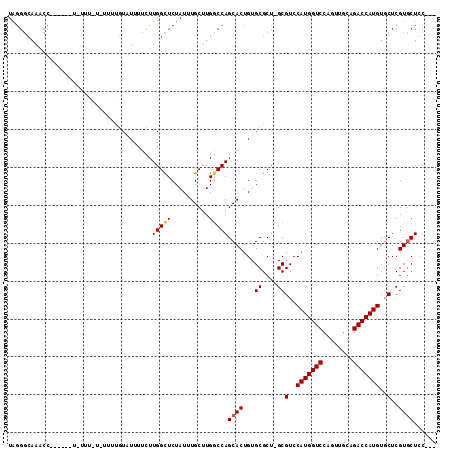
>X_DroMel_CAF1 8778359 104 + 22224390 UAGGGCAAGCCC-----UAUUUCUAUUUUGUAUUUUCUUGGCUCUAUUUGCUUGGCCAGCACUGUGCGCU-GCGUCCAUGGUCCAGUUGCAGACCAUGUGCUCGUCCUGC--- ((((((.(((..-----............((((....((((((..........))))))....))))...-.....(((((((........))))))).))).)))))).--- ( -29.10) >DroSec_CAF1 26926 109 + 1 UGGGGCCAACCAUUACUUUUUUUUUUUUUGUAUUUUCUUGGCUCUAUUUGCAUGCCCAGCACUGUGCGCU-GCGUCCAUGGUCCAGUUGCAGACCAUGUGCUCGUGCUCC--- (((((((((....(((.............))).....)))))))))...(((((..((((.......)))-).(..(((((((........)))))))..).)))))...--- ( -32.92) >DroYak_CAF1 23593 93 + 1 UAAGUGAAA--------------------CUGUUUUCUUGGUUCUAUUUGCUUGGCCAGCACUGUGCGCUGGCGUCCAUGGUCCAGUUGCAGACCAUGUUCUCGUGCUCCUGC .(((((.((--------------------(((......))))).)))))((.((((((((.......))))))(..(((((((........)))))))..).)).))...... ( -27.30) >consensus UAGGGCAAACC______U_UUU_U_UUUUGUAUUUUCUUGGCUCUAUUUGCUUGGCCAGCACUGUGCGCU_GCGUCCAUGGUCCAGUUGCAGACCAUGUGCUCGUGCUCC___ ......................................(((((..........)))))((((...((....))(..(((((((........)))))))..)..))))...... (-18.08 = -18.63 + 0.56)

| Location | 8,778,359 – 8,778,463 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.76 |
| Mean single sequence MFE | -31.83 |
| Consensus MFE | -19.09 |
| Energy contribution | -19.53 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.60 |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
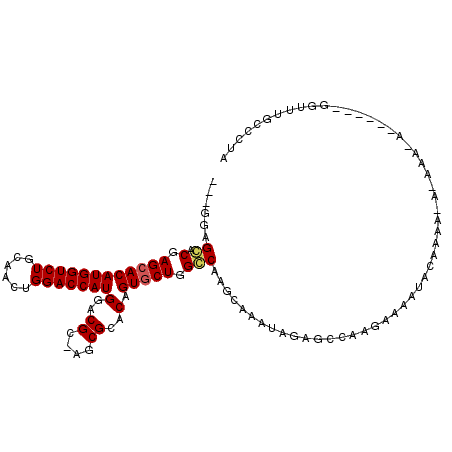
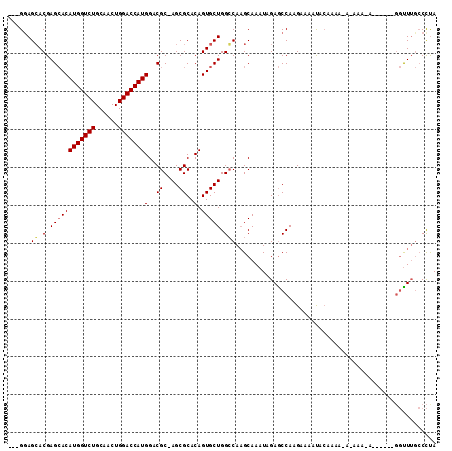
>X_DroMel_CAF1 8778359 104 - 22224390 ---GCAGGACGAGCACAUGGUCUGCAACUGGACCAUGGACGC-AGCGCACAGUGCUGGCCAAGCAAAUAGAGCCAAGAAAAUACAAAAUAGAAAUA-----GGGCUUGCCCUA ---..(((.(((((.((((((((......))))))))....(-(((((...)))))).((..((.......))...............((....))-----))))))).))). ( -31.60) >DroSec_CAF1 26926 109 - 1 ---GGAGCACGAGCACAUGGUCUGCAACUGGACCAUGGACGC-AGCGCACAGUGCUGGGCAUGCAAAUAGAGCCAAGAAAAUACAAAAAAAAAAAAAGUAAUGGUUGGCCCCA ---((.((.(((.((((((((((......)))))))).((.(-(((((...))))))(((...........))).......................))..)).))))).)). ( -32.20) >DroYak_CAF1 23593 93 - 1 GCAGGAGCACGAGAACAUGGUCUGCAACUGGACCAUGGACGCCAGCGCACAGUGCUGGCCAAGCAAAUAGAACCAAGAAAACAG--------------------UUUCACUUA ...(((((....(..((((((((......))))))))..)((((((((...))))))))........................)--------------------))))..... ( -31.70) >consensus ___GGAGCACGAGCACAUGGUCUGCAACUGGACCAUGGACGC_AGCGCACAGUGCUGGCCAAGCAAAUAGAGCCAAGAAAAUACAAAA_A_AAA_A______GGUUUGCCCUA ......((.(.((((((((((((......)))))))(..((....))..).))))).)))..................................................... (-19.09 = -19.53 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:49 2006