
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,759,300 – 8,759,414 |
| Length | 114 |
| Max. P | 0.999998 |

| Location | 8,759,300 – 8,759,414 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 72.22 |
| Mean single sequence MFE | -31.33 |
| Consensus MFE | -21.39 |
| Energy contribution | -23.39 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -4.41 |
| Structure conservation index | 0.68 |
| SVM decision value | 6.19 |
| SVM RNA-class probability | 0.999997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8759300 114 + 22224390 -CCAAAAGUAUGCAGCGAACGUGAAAAAUGCCGUUUUCCGUUUUCCGCUGCAUUCUUUUGCACUUUCGCACACUAGUAUUGUUGAUAAUCCAAGGUAUUUUUGUGUCAC-CACCUU -.((((((.((((((((((((.(((((......)))))))))...)))))))).)))))).......(((((..((((((.(((......)))))))))..)))))...-...... ( -32.70) >DroSec_CAF1 7394 93 + 1 CCCAAAAGUAUGCAGCAA-------------UGUUUUCCAU--UCUGCUGCAUACUUUUG--------CACACUAGAAUUGUGGAUUUUCCAAGAGUUUUUCGUGGCACACGCCUU ..((((((((((((((((-------------((.....)))--..)))))))))))))))--------.((...((((((.(((.....)))..))))))..))(((....))).. ( -30.80) >DroSim_CAF1 8367 88 + 1 -----AGGUAUGCAGCAA-------------UGUUUUCCAU--UCUGCUGCAUACUUUUG--------CACACUAGAAUUGUGGAUUUUCCAAGAGAUUUUCGUGGCACACACCUU -----(((((((((((((-------------((.....)))--..)))))))))))).((--------(.(((.((((((.(((.....)))...)))))).))))))........ ( -30.50) >consensus _CCAAAAGUAUGCAGCAA_____________UGUUUUCCAU__UCUGCUGCAUACUUUUG________CACACUAGAAUUGUGGAUUUUCCAAGAGAUUUUCGUGGCACACACCUU ..(((((((((((((((............................)))))))))))))))..........(((.((((((.(((.....)))...)))))).)))........... (-21.39 = -23.39 + 2.00)

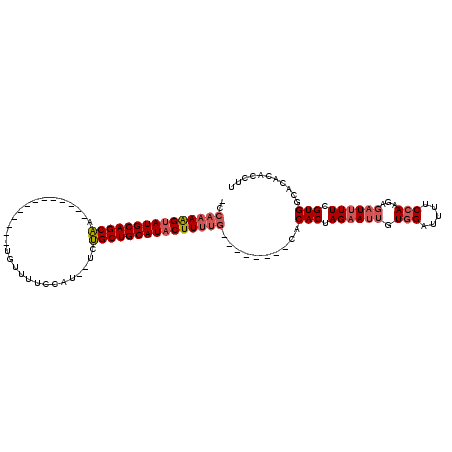

| Location | 8,759,300 – 8,759,414 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 72.22 |
| Mean single sequence MFE | -30.19 |
| Consensus MFE | -20.27 |
| Energy contribution | -21.17 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.67 |
| SVM decision value | 6.30 |
| SVM RNA-class probability | 0.999998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8759300 114 - 22224390 AAGGUG-GUGACACAAAAAUACCUUGGAUUAUCAACAAUACUAGUGUGCGAAAGUGCAAAAGAAUGCAGCGGAAAACGGAAAACGGCAUUUUUCACGUUCGCUGCAUACUUUUGG- ...(((-....))).......((......................(..(....)..)(((((.(((((((((...((((((((......))))).)))))))))))).)))))))- ( -34.50) >DroSec_CAF1 7394 93 - 1 AAGGCGUGUGCCACGAAAAACUCUUGGAAAAUCCACAAUUCUAGUGUG--------CAAAAGUAUGCAGCAGA--AUGGAAAACA-------------UUGCUGCAUACUUUUGGG ..(((....)))(((.........(((.....))).........))).--------(((((((((((((((..--(((.....))-------------)))))))))))))))).. ( -31.07) >DroSim_CAF1 8367 88 - 1 AAGGUGUGUGCCACGAAAAUCUCUUGGAAAAUCCACAAUUCUAGUGUG--------CAAAAGUAUGCAGCAGA--AUGGAAAACA-------------UUGCUGCAUACCU----- ........((((((((.....)).(((.....)))........))).)--------))...((((((((((..--(((.....))-------------)))))))))))..----- ( -25.00) >consensus AAGGUGUGUGCCACGAAAAUCUCUUGGAAAAUCCACAAUUCUAGUGUG________CAAAAGUAUGCAGCAGA__AUGGAAAACA_____________UUGCUGCAUACUUUUGG_ ...........((((.........(((.....))).........))))........((((((((((((((((....((.....)).............)))))))))))))))).. (-20.27 = -21.17 + 0.89)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:43 2006