
| Sequence ID | X_DroMel_CAF1 |
|---|---|
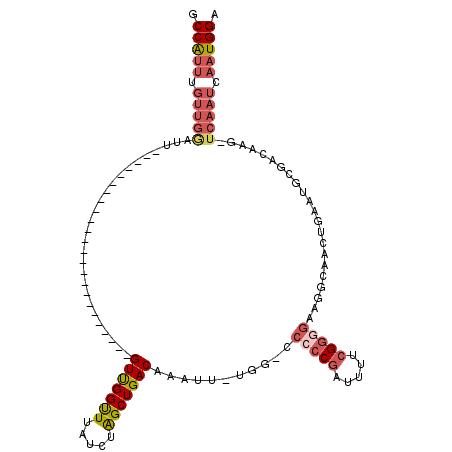
| Location | 8,729,904 – 8,729,996 |
| Length | 92 |
| Max. P | 0.996597 |

| Location | 8,729,904 – 8,729,996 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.73 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -18.26 |
| Energy contribution | -19.60 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.996597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
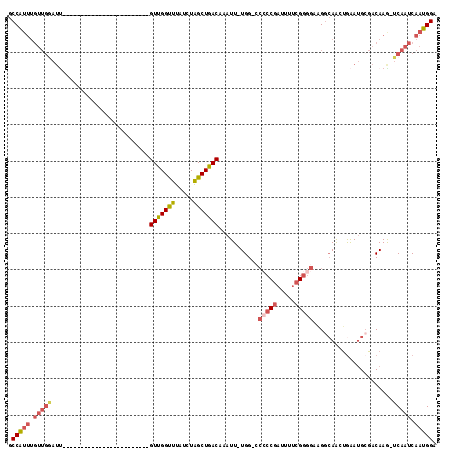
>X_DroMel_CAF1 8729904 92 + 22224390 GCCAUUUGUUGGAUU------------------------GUUGGUUUAUCUAGCUGACAAAUUUUGG-GCAACGAUUUUCGGGGAAGGCAACUGAAUGCGACAAG-UCAAUCAAUGGA .(((((.(((((.((------------------------((..(((.....)))..))))..(((((-....).....(((.(..((....))...).)))))))-))))).))))). ( -24.40) >DroSec_CAF1 10322 91 + 1 GCCAUUUGUUGGAUU------------------------GUCGGUUUAUCUAGCUGACAAAUU-UGG-CCCCCGAUUUUCGGGGAAGGCAACUGAAUGCGACAAA-UCAAUCAAUGGA .(((((.(((((.((------------------------(((((((.....))))))))).((-((.-.(((((.....)))))...(((......)))..))))-))))).))))). ( -32.70) >DroSim_CAF1 10802 91 + 1 GCCAUUUGUUGGAUU------------------------GUUGGUUUAUCUAGCUGACAAAUU-UGG-CCCCCGAUUUUCGGGGAAGGCAACUGAAUGCGACAAG-UCAAUCAAUGGA .(((((.((((..((------------------------((((((((.....((..(.....)-..)-)(((((.....))))).((....)))))).)))))).-.)))).))))). ( -30.70) >DroEre_CAF1 10465 99 + 1 GCCAUUUGUUGAAUCG----------------UUGGUUUGUUGGCUUAUCUAGCUGACAAAUU-UGG-CCCCCGAUUUUCGGGGAAGGCAAAUAAAUGCGACAAG-UCAAUCAAUGGA .(((((.(((((...(----------------(((..((((..(((.....)))..))))(((-((.-((((((.....)))))..).))))).....))))...-))))).))))). ( -37.50) >DroYak_CAF1 10833 116 + 1 GCCAUUUGUUGGAUUGUUGCUUUGUUGCUUUGUUGGUUUGUUGGUUUAUCUAGCUGACAAAUU-UGGUCCCCCGAUUUUGGGGGACGGCAAGUGAAUGAGACAAG-UCAAUCAAUGGA .(((((.((((..(((((......(..(((.((((((((((..(((.....)))..)))))))-..((((((((....))))))))))))))..)....))))).-.)))).))))). ( -44.60) >DroAna_CAF1 8857 79 + 1 GCCG--------------------------------UGUGUUGGUUUAUCUGGCUGACAAAUU-UGG----CCGAUUGCCGGGGGCG--AGGGGGAAGCCCCAAGCUCAAUCAGUGGA ((((--------------------------------..(((..(((.....)))..)))....-)))----)(.((((....((((.--.((((....))))..))))...)))).). ( -31.10) >consensus GCCAUUUGUUGGAUU________________________GUUGGUUUAUCUAGCUGACAAAUU_UGG_CCCCCGAUUUUCGGGGAAGGCAACUGAAUGCGACAAG_UCAAUCAAUGGA .(((((.(((((...........................(((((((.....)))))))...........(((((.....)))))......................))))).))))). (-18.26 = -19.60 + 1.34)

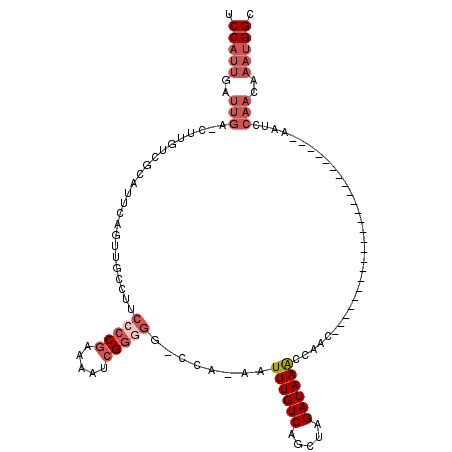
| Location | 8,729,904 – 8,729,996 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.73 |
| Mean single sequence MFE | -24.08 |
| Consensus MFE | -11.49 |
| Energy contribution | -13.18 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
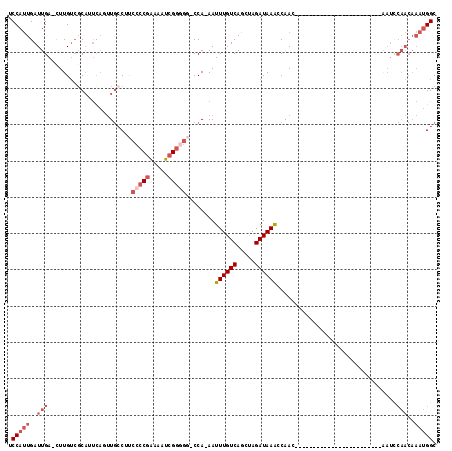
>X_DroMel_CAF1 8729904 92 - 22224390 UCCAUUGAUUGA-CUUGUCGCAUUCAGUUGCCUUCCCCGAAAAUCGUUGC-CCAAAAUUUGUCAGCUAGAUAAACCAAC------------------------AAUCCAACAAAUGGC .(((((..(((.-.((((.(((......)))...................-......((((((.....))))))...))------------------------))..)))..))))). ( -13.50) >DroSec_CAF1 10322 91 - 1 UCCAUUGAUUGA-UUUGUCGCAUUCAGUUGCCUUCCCCGAAAAUCGGGGG-CCA-AAUUUGUCAGCUAGAUAAACCGAC------------------------AAUCCAACAAAUGGC .(((((..(((.-.((((((((......)))..((((((.....))))))-...-..((((((.....))))))..)))------------------------))..)))..))))). ( -28.90) >DroSim_CAF1 10802 91 - 1 UCCAUUGAUUGA-CUUGUCGCAUUCAGUUGCCUUCCCCGAAAAUCGGGGG-CCA-AAUUUGUCAGCUAGAUAAACCAAC------------------------AAUCCAACAAAUGGC .(((((..(((.-.((((.(((......)))..((((((.....))))))-...-..((((((.....))))))...))------------------------))..)))..))))). ( -23.60) >DroEre_CAF1 10465 99 - 1 UCCAUUGAUUGA-CUUGUCGCAUUUAUUUGCCUUCCCCGAAAAUCGGGGG-CCA-AAUUUGUCAGCUAGAUAAGCCAACAAACCAA----------------CGAUUCAACAAAUGGC .(((((..((((-.((((.(((......)))..((((((.....))))))-...-..(((((..(((.....)))..)))))...)----------------))).))))..))))). ( -28.00) >DroYak_CAF1 10833 116 - 1 UCCAUUGAUUGA-CUUGUCUCAUUCACUUGCCGUCCCCCAAAAUCGGGGGACCA-AAUUUGUCAGCUAGAUAAACCAACAAACCAACAAAGCAACAAAGCAACAAUCCAACAAAUGGC .(((((..(((.-.((((.........((((.(((((((......)))))))..-..((((((.....))))))................)))).......))))..)))..))))). ( -28.29) >DroAna_CAF1 8857 79 - 1 UCCACUGAUUGAGCUUGGGGCUUCCCCCU--CGCCCCCGGCAAUCGG----CCA-AAUUUGUCAGCCAGAUAAACCAACACA--------------------------------CGGC .((.((((((..(((.(((((........--.))))).)))))))))----...-..((((((.....))))))........--------------------------------.)). ( -22.20) >consensus UCCAUUGAUUGA_CUUGUCGCAUUCAGUUGCCUUCCCCGAAAAUCGGGGG_CCA_AAUUUGUCAGCUAGAUAAACCAAC________________________AAUCCAACAAAUGGC .(((((..(((.......................(((((.....)))))........((((((.....)))))).................................)))..))))). (-11.49 = -13.18 + 1.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:35 2006