
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,700,749 – 8,700,841 |
| Length | 92 |
| Max. P | 0.743068 |

| Location | 8,700,749 – 8,700,841 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 91.77 |
| Mean single sequence MFE | -29.74 |
| Consensus MFE | -24.66 |
| Energy contribution | -24.82 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
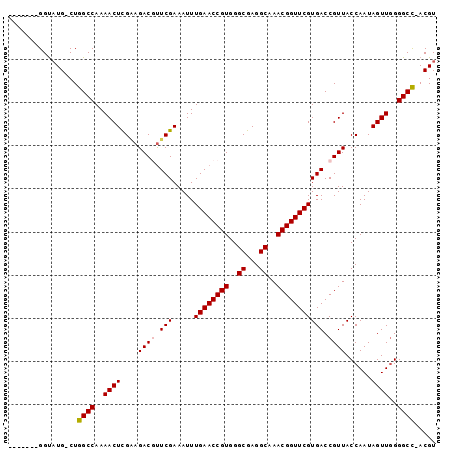
>X_DroMel_CAF1 8700749 92 + 22224390 -------GGUAUGGCUGGCCAAAACUCGAAGACGUUCGAAAUUUGAACCGUGGGCGAGGCAAACGGUUCGUGACCGUUACCAAUAGUUGGGGCCAACGU -------...(((..(((((..((((....((((.(((.....((((((((..((...))..))))))))))).))))......))))..))))).))) ( -29.60) >DroSec_CAF1 136149 90 + 1 -------GGUAUG-CUGGCCAAAACUCGAAGACAUUCGAAAUUUGAACCGUGGGCGAGGCAAACGGUUCGUGACCGUUACCAAUAGUUGGGGCU-ACGU -------...(((-.(((((..((((((((....))))).....(((((((..((...))..)))))))................)))..))))-)))) ( -27.40) >DroSim_CAF1 101080 90 + 1 -------GGUAUG-CUGGCCAAAACUCGAAGACAUUCGAAAUUUGAACCGUGGGCGAGGCAAACGGUUCGUGACCGUUACCAAUAGUUGGGGCU-ACGU -------...(((-.(((((..((((((((....))))).....(((((((..((...))..)))))))................)))..))))-)))) ( -27.40) >DroEre_CAF1 150622 90 + 1 -------GGCUUG-CUGGCCAAAACUUGGAGACGUUCGAAAUUUGAACCGUGGGCGAGGCAAACGGUUCGUGACCGUUACCAAUAGUUUGGGCC-ACGU -------.....(-((((((.((((((((.((((.(((.....((((((((..((...))..))))))))))).)))).)))..))))).))))-).)) ( -34.20) >DroYak_CAF1 142649 97 + 1 UAUAUAUGGUAUG-CUGGCCAAAACUUGCAGACGCUCGAAAUUUGAACCGUGGGCGAGGCAAACGGUUCGUGACCGUUACCAAUAGUUGGGGCC-ACGU ..........(((-.(((((..((((((..((((.(((.....((((((((..((...))..))))))))))).))))..))..))))..))))-)))) ( -30.10) >consensus _______GGUAUG_CUGGCCAAAACUCGAAGACGUUCGAAAUUUGAACCGUGGGCGAGGCAAACGGUUCGUGACCGUUACCAAUAGUUGGGGCC_ACGU ................((((..((((....((((.(((.....((((((((..((...))..))))))))))).))))......))))..))))..... (-24.66 = -24.82 + 0.16)


| Location | 8,700,749 – 8,700,841 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 91.77 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -20.42 |
| Energy contribution | -20.42 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.743068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
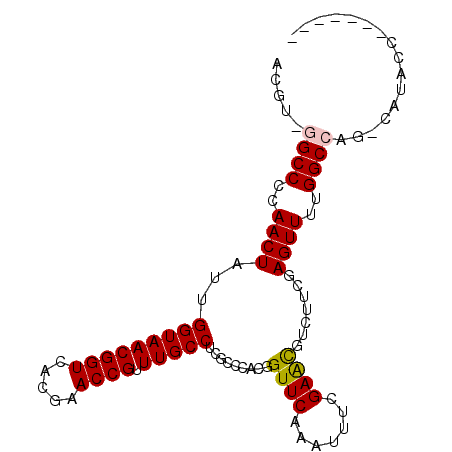
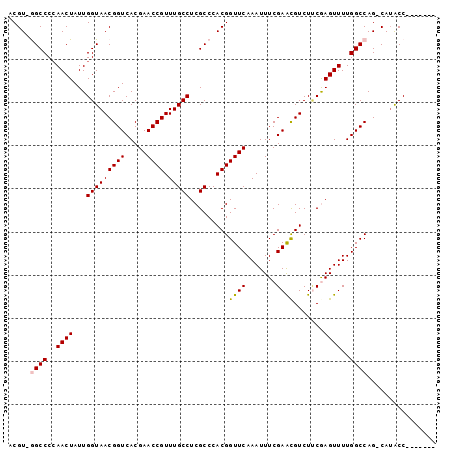
>X_DroMel_CAF1 8700749 92 - 22224390 ACGUUGGCCCCAACUAUUGGUAACGGUCACGAACCGUUUGCCUCGCCCACGGUUCAAAUUUCGAACGUCUUCGAGUUUUGGCCAGCCAUACC------- ..((((....))))...((((...(((((.(((((((..((...))..)))))))....((((((....))))))...))))).))))....------- ( -28.40) >DroSec_CAF1 136149 90 - 1 ACGU-AGCCCCAACUAUUGGUAACGGUCACGAACCGUUUGCCUCGCCCACGGUUCAAAUUUCGAAUGUCUUCGAGUUUUGGCCAG-CAUACC------- ..((-(((.(((.....)))....(((((.(((((((..((...))..)))))))....((((((....))))))...))))).)-).))).------- ( -25.40) >DroSim_CAF1 101080 90 - 1 ACGU-AGCCCCAACUAUUGGUAACGGUCACGAACCGUUUGCCUCGCCCACGGUUCAAAUUUCGAAUGUCUUCGAGUUUUGGCCAG-CAUACC------- ..((-(((.(((.....)))....(((((.(((((((..((...))..)))))))....((((((....))))))...))))).)-).))).------- ( -25.40) >DroEre_CAF1 150622 90 - 1 ACGU-GGCCCAAACUAUUGGUAACGGUCACGAACCGUUUGCCUCGCCCACGGUUCAAAUUUCGAACGUCUCCAAGUUUUGGCCAG-CAAGCC------- ...(-((((.(((((..(((..(((.((..(((((((..((...))..))))))).......)).)))..)))))))).))))).-......------- ( -28.50) >DroYak_CAF1 142649 97 - 1 ACGU-GGCCCCAACUAUUGGUAACGGUCACGAACCGUUUGCCUCGCCCACGGUUCAAAUUUCGAGCGUCUGCAAGUUUUGGCCAG-CAUACCAUAUAUA ...(-((((..((((...(((((((((.....)))).)))))..((..(((.(((.......))))))..)).))))..))))).-............. ( -26.60) >consensus ACGU_GGCCCCAACUAUUGGUAACGGUCACGAACCGUUUGCCUCGCCCACGGUUCAAAUUUCGAACGUCUUCGAGUUUUGGCCAG_CAUACC_______ .....((((..((((...(((((((((.....)))).))))).........((((.......)))).......))))..))))................ (-20.42 = -20.42 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:23 2006