
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,679,274 – 8,679,396 |
| Length | 122 |
| Max. P | 0.980147 |

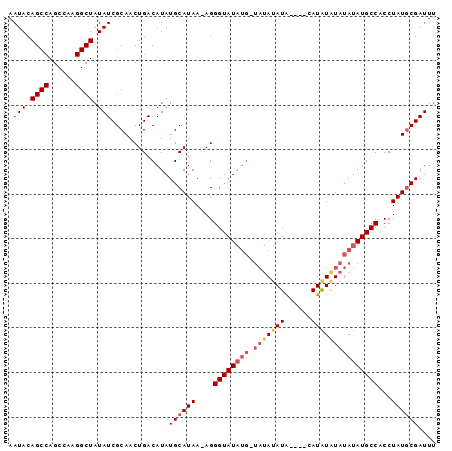
| Location | 8,679,274 – 8,679,370 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 86.22 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -16.15 |
| Energy contribution | -18.60 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8679274 96 + 22224390 AAUACAGCCAGCCAAGGCUAUAUCGCAACUGACAUAUGCAUAAAAGGGUAUAUGAUAUAUAUGUAAAUAUGUGUAUAUAUGCCACCUAUGCGAUUG .(((.((((......)))).))).............((((((....((((((((((((((((....)))))))).))))))))...)))))).... ( -26.00) >DroSec_CAF1 115761 84 + 1 AAUACAGCCAGCCAAGGCUAUAUCGCAACUGACAUAUGCAUAA-AGGGUAU-------ACAUA----CAUAUAUAUAUAUGCCACCUAUACGAUUU .....((((......))))..((((((.........)))....-..(((((-------(.(((----......))).))))))........))).. ( -13.80) >DroSim_CAF1 80488 91 + 1 AAUACAGCCAGCCAAGGCUAUAUCGCAACUGACAUAUGCAUAA-AGGGUAUAUGUUAUAUAUA----CAUAUAUAUAUAUGCCACCUAUGCGAUUU .(((.((((......)))).))).............((((((.-..(((((((((.(((((..----..))))))))))))))...)))))).... ( -23.50) >consensus AAUACAGCCAGCCAAGGCUAUAUCGCAACUGACAUAUGCAUAA_AGGGUAUAUG_UAUAUAUA____CAUAUAUAUAUAUGCCACCUAUGCGAUUU .(((.((((......)))).))).............((((((....((((((((.(((((((......)))))))))))))))...)))))).... (-16.15 = -18.60 + 2.45)


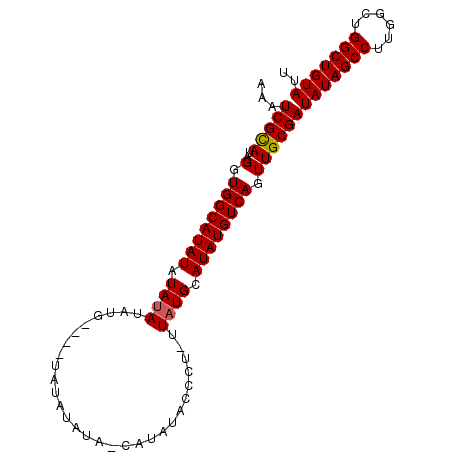
| Location | 8,679,274 – 8,679,370 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 86.22 |
| Mean single sequence MFE | -26.13 |
| Consensus MFE | -21.77 |
| Energy contribution | -21.88 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.60 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.980147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8679274 96 - 22224390 CAAUCGCAUAGGUGGCAUAUAUACACAUAUUUACAUAUAUAUCAUAUACCCUUUUAUGCAUAUGUCAGUUGCGAUAUAGCCUUGGCUGGCUGUAUU ...(((((..(.((((((((......((((......))))..((((........)))).)))))))).))))))(((((((......))))))).. ( -24.30) >DroSec_CAF1 115761 84 - 1 AAAUCGUAUAGGUGGCAUAUAUAUAUAUG----UAUGU-------AUACCCU-UUAUGCAUAUGUCAGUUGCGAUAUAGCCUUGGCUGGCUGUAUU .....(((((((.((.((((((((....)----)))))-------)).)).)-)))))).............(((((((((......))))))))) ( -24.90) >DroSim_CAF1 80488 91 - 1 AAAUCGCAUAGGUGGCAUAUAUAUAUAUG----UAUAUAUAACAUAUACCCU-UUAUGCAUAUGUCAGUUGCGAUAUAGCCUUGGCUGGCUGUAUU ...(((((..(.((((((((.((((...(----(((((.....))))))...-.)))).)))))))).))))))(((((((......))))))).. ( -29.20) >consensus AAAUCGCAUAGGUGGCAUAUAUAUAUAUG____UAUAUAUA_CAUAUACCCU_UUAUGCAUAUGUCAGUUGCGAUAUAGCCUUGGCUGGCUGUAUU ...(((((..(.((((((((.((((.............................)))).)))))))).))))))(((((((......))))))).. (-21.77 = -21.88 + 0.11)

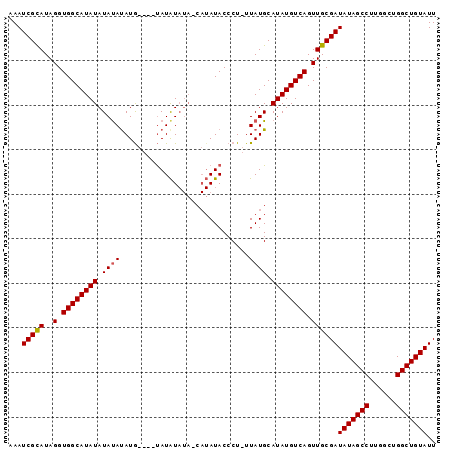
| Location | 8,679,295 – 8,679,396 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 84.90 |
| Mean single sequence MFE | -19.89 |
| Consensus MFE | -13.09 |
| Energy contribution | -15.53 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.769671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8679295 101 + 22224390 AUCGCAACUGACAUAUGCAUAAAAGGGUAUAUGAUAUAUAUGUAAAUAUGUGUAUAUAUGCCACCUAUGCGAUUGCAAGACCCUUCCACACGCCUGUCACU ........(((((..((((((....((((((((((((((((....)))))))).))))))))...))))))...((...............)).))))).. ( -23.76) >DroSec_CAF1 115782 89 + 1 AUCGCAACUGACAUAUGCAUAA-AGGGUAU-------ACAUA----CAUAUAUAUAUAUGCCACCUAUACGAUUUCGAAACCCUUCCUCACGCCUGUCACU ........(((((..((....(-(((((..-------.....----(((((....))))).........((....))..)))))).....))..))))).. ( -14.10) >DroSim_CAF1 80509 96 + 1 AUCGCAACUGACAUAUGCAUAA-AGGGUAUAUGUUAUAUAUA----CAUAUAUAUAUAUGCCACCUAUGCGAUUUCGAAACCCUUCCUCACGCCUGUCACU ........(((((..((((((.-..(((((((((.(((((..----..))))))))))))))...)))))).....(((....)))........))))).. ( -21.80) >consensus AUCGCAACUGACAUAUGCAUAA_AGGGUAUAUG_UAUAUAUA____CAUAUAUAUAUAUGCCACCUAUGCGAUUUCGAAACCCUUCCUCACGCCUGUCACU ........(((((...(((((....((((((((.(((((((......)))))))))))))))...)))))(....)..................))))).. (-13.09 = -15.53 + 2.45)

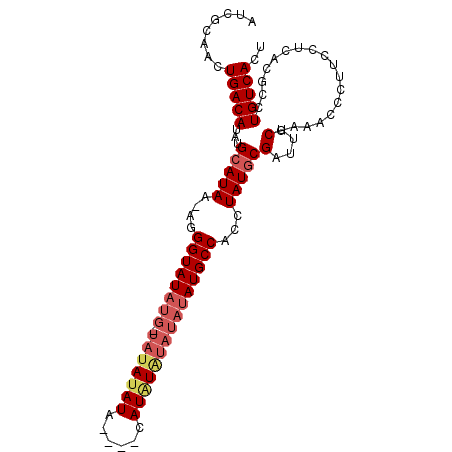

| Location | 8,679,295 – 8,679,396 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 84.90 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -19.22 |
| Energy contribution | -21.00 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8679295 101 - 22224390 AGUGACAGGCGUGUGGAAGGGUCUUGCAAUCGCAUAGGUGGCAUAUAUACACAUAUUUACAUAUAUAUCAUAUACCCUUUUAUGCAUAUGUCAGUUGCGAU .(..((.((((((((((((((((((((....))).))(((..((((((............))))))..)))..)))))))....)))))))).))..)... ( -28.60) >DroSec_CAF1 115782 89 - 1 AGUGACAGGCGUGAGGAAGGGUUUCGAAAUCGUAUAGGUGGCAUAUAUAUAUAUG----UAUGU-------AUACCCU-UUAUGCAUAUGUCAGUUGCGAU ..(((((.((((((((...((((....)))).....((((((((((((....)))----)))))-------.))))))-))))))...)))))........ ( -25.90) >DroSim_CAF1 80509 96 - 1 AGUGACAGGCGUGAGGAAGGGUUUCGAAAUCGCAUAGGUGGCAUAUAUAUAUAUG----UAUAUAUAACAUAUACCCU-UUAUGCAUAUGUCAGUUGCGAU .(..((.((((((..(((((((..((....)).....(((..((((((((....)----)))))))..)))..)))))-)).....)))))).))..)... ( -25.90) >consensus AGUGACAGGCGUGAGGAAGGGUUUCGAAAUCGCAUAGGUGGCAUAUAUAUAUAUG____UAUAUAUA_CAUAUACCCU_UUAUGCAUAUGUCAGUUGCGAU ..(((((.((((((...(((((...(..(((.....)))..).((((((((........))))))))......))))).))))))...)))))........ (-19.22 = -21.00 + 1.78)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:17 2006