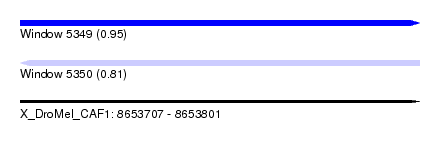
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,653,707 – 8,653,801 |
| Length | 94 |
| Max. P | 0.947548 |

| Location | 8,653,707 – 8,653,801 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 82.24 |
| Mean single sequence MFE | -38.27 |
| Consensus MFE | -27.83 |
| Energy contribution | -28.30 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.68 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
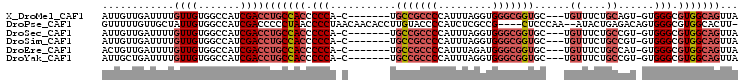
>X_DroMel_CAF1 8653707 94 + 22224390 UAACUGCCACGCCCAC-ACUGCAGAAACA---GCACCGCCCACCUAAAUGGGGCGGCA-------G-UGGGGGUGGCAGGUCGAUGGCCACAACAAAAUCAACAAU ...(((((((.(((((-..(((.......---)))((((((.........))))))..-------)-)))).)))))))((.(((............))).))... ( -40.70) >DroPse_CAF1 119970 99 + 1 -AAGUGCCACGCCCACUGUCUCAGUAU--UUGGGAG----CGGCGAGAUGGGGUACAAGGUGUUGUUAGGGGUUAGGGGGUCGAUGGCCACAAUAGCAACAAAAAC -..(((((.(((.(.(((((((((...--)))))).----))).).).)).)))))....((((((((..(((((.........)))))....))))))))..... ( -27.80) >DroSec_CAF1 90847 94 + 1 UAACUGCCACGCCCAC-ACGGCAGAAACA---GCACCGCCCACCUAAAUGGGGCGGCA-------G-UGGGGGUGGCAGGUCGAUGGCCACAACAAAAUCAACAAU ...(((((((.(((((-...((.......---)).((((((.........))))))..-------)-)))).)))))))((.(((............))).))... ( -40.20) >DroSim_CAF1 54763 94 + 1 UAACUGCCACGCCCAC-ACGGCAGAAACA---GCACCGCCCACCUAAAUGGGGCGGCA-------G-UGGGGGUGGCAGGUCGAUGGCCACAACAAAAUCAACAAU ...(((((((.(((((-...((.......---)).((((((.........))))))..-------)-)))).)))))))((.(((............))).))... ( -40.20) >DroEre_CAF1 102830 94 + 1 UAACUGCCACGCCCAC-AUGGCAGAAACA---GCACCGCCCAUCUAAAUGGGGCGGCA-------G-UGGGGGUGGCAGGUCGAUGGCCACAACAAAAUCAACAGU ...(((((((.(((((-.((((.......---)).((((((.........))))))))-------)-)))).)))))))((.(((............))).))... ( -40.20) >DroYak_CAF1 93671 94 + 1 UAACUGCCACGCCCAC-ACGGCAGAAACA---GCACCGCCCACCUAAAUGGGGCGGCA-------G-UGGGGGUGGCAGGUCGAUGGCCACAACAAAAUCAGCAAU ...(((((((.(((((-...((.......---)).((((((.........))))))..-------)-)))).)))))))((.(((............))).))... ( -40.50) >consensus UAACUGCCACGCCCAC_ACGGCAGAAACA___GCACCGCCCACCUAAAUGGGGCGGCA_______G_UGGGGGUGGCAGGUCGAUGGCCACAACAAAAUCAACAAU ....((((((.(((......((..........)).((((((.........))))))............))).))))))(((.....)))................. (-27.83 = -28.30 + 0.47)


| Location | 8,653,707 – 8,653,801 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 82.24 |
| Mean single sequence MFE | -37.53 |
| Consensus MFE | -23.72 |
| Energy contribution | -25.30 |
| Covariance contribution | 1.58 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
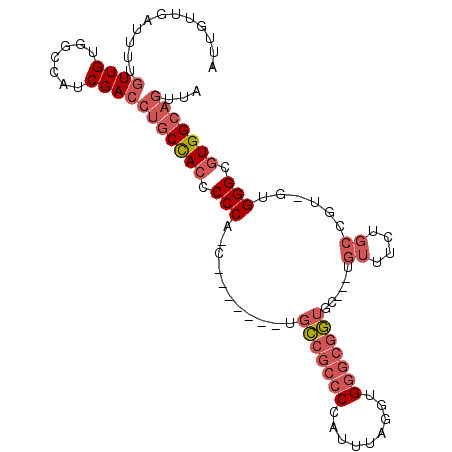
>X_DroMel_CAF1 8653707 94 - 22224390 AUUGUUGAUUUUGUUGUGGCCAUCGACCUGCCACCCCCA-C-------UGCCGCCCCAUUUAGGUGGGCGGUGC---UGUUUCUGCAGU-GUGGGCGUGGCAGUUA ...((((((...((....)).))))))(((((((.((((-(-------.((((((((......).)))))))((---(((....)))))-))))).)))))))... ( -45.30) >DroPse_CAF1 119970 99 - 1 GUUUUUGUUGCUAUUGUGGCCAUCGACCCCCUAACCCCUAACAACACCUUGUACCCCAUCUCGCCG----CUCCCAA--AUACUGAGACAGUGGGCGUGGCACUU- (((..(((..(....)..).))..))).............((((....))))..........((((----(.(((..--..((((...))))))).)))))....- ( -19.80) >DroSec_CAF1 90847 94 - 1 AUUGUUGAUUUUGUUGUGGCCAUCGACCUGCCACCCCCA-C-------UGCCGCCCCAUUUAGGUGGGCGGUGC---UGUUUCUGCCGU-GUGGGCGUGGCAGUUA ...((((((...((....)).))))))(((((((.((((-(-------.((((((((......).)))))))((---.......))...-))))).)))))))... ( -40.80) >DroSim_CAF1 54763 94 - 1 AUUGUUGAUUUUGUUGUGGCCAUCGACCUGCCACCCCCA-C-------UGCCGCCCCAUUUAGGUGGGCGGUGC---UGUUUCUGCCGU-GUGGGCGUGGCAGUUA ...((((((...((....)).))))))(((((((.((((-(-------.((((((((......).)))))))((---.......))...-))))).)))))))... ( -40.80) >DroEre_CAF1 102830 94 - 1 ACUGUUGAUUUUGUUGUGGCCAUCGACCUGCCACCCCCA-C-------UGCCGCCCCAUUUAGAUGGGCGGUGC---UGUUUCUGCCAU-GUGGGCGUGGCAGUUA ...((((((...((....)).))))))(((((((.((((-(-------.(((((((.........)))))))((---.......))...-))))).)))))))... ( -39.70) >DroYak_CAF1 93671 94 - 1 AUUGCUGAUUUUGUUGUGGCCAUCGACCUGCCACCCCCA-C-------UGCCGCCCCAUUUAGGUGGGCGGUGC---UGUUUCUGCCGU-GUGGGCGUGGCAGUUA ............((((.......))))(((((((.((((-(-------.((((((((......).)))))))((---.......))...-))))).)))))))... ( -38.80) >consensus AUUGUUGAUUUUGUUGUGGCCAUCGACCUGCCACCCCCA_C_______UGCCGCCCCAUUUAGGUGGGCGGUGC___UGUUUCUGCCGU_GUGGGCGUGGCAGUUA ............((((.......))))(((((((.(((...........(((((((.........)))))))......((....))......))).)))))))... (-23.72 = -25.30 + 1.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:08 2006