
| Sequence ID | X_DroMel_CAF1 |
|---|---|
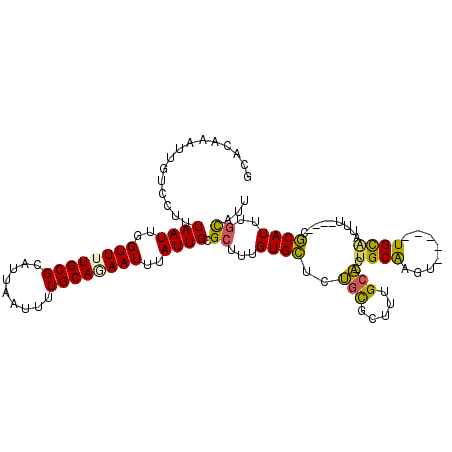
| Location | 8,642,724 – 8,642,836 |
| Length | 112 |
| Max. P | 0.977336 |

| Location | 8,642,724 – 8,642,836 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.33 |
| Mean single sequence MFE | -26.41 |
| Consensus MFE | -12.89 |
| Energy contribution | -12.05 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8642724 112 + 22224390 GCACAAAUUGUCCUUUAAUUGGUUUUGCGCAUUAAUUUUGCAAAAUUUAUUGCGCUUUGUGCUCGCCGCUUUGCC-GUGCAAGUCCGGCUGCAUUUUCAUUCGCAUUUGCAUU (((((((....((.......))....(((((..(((((....)))))...))))))))))))..((((.(((((.-..)))))..))))((((..............)))).. ( -28.44) >DroPse_CAF1 108446 103 + 1 G-CGAAAUUGUCCUUUAAUUGGUUUUGCGCAUUAAUUUUGCACAAUUUAUUGCGCUUUGUGUGUUGUGCUGUGCAUCUGCAAGU-----UGCAACUU---GCACAUUUGUG-U (-((((((((.........)))))))))((((.......(((((((.(((........))).)))))))((((((..(((....-----.)))...)---)))))...)))-) ( -26.90) >DroSim_CAF1 43665 112 + 1 GCACAAAUUGUCCUUUAAUUGGUUUUGCGCAUUAAUUUUGCAGAAUUUAUUGCGCUUUGUGCUCGGCGCUUUGCC-GUGCGAGUCCUGCUGCAUUUUCAUUCGCAUUCGCAUU (((((((....((.......))....(((((..(((((....)))))...)))))))))))).((((.....)))-)((((((...(((.............))))))))).. ( -30.32) >DroEre_CAF1 92020 88 + 1 CCACAAAUUGUCCUUUAAUUGGUUUUGCGCAUUAAUUUUGCAGAAUUUAUUGCGCUUUGUGCUCUGCGUUUUGCA-CU------------------------GCAUUUGCAUU .((((((....((.......))....(((((..(((((....)))))...)))))))))))...((((...(((.-..------------------------)))..)))).. ( -18.60) >DroYak_CAF1 82457 105 + 1 CCACAAAUUGUCCUUUAAUUGGUUUUGCGCAUUAAUUUUGCAGAAUUUAUUGCGCUUUGUGCUUUGCGUUUUGCA-UUGCGAGUUCGGUUGUGGU-------GCAUUUUCAUU ((((((.(((..(((...........(((((..(((((....)))))...))))).....((..(((.....)))-..)))))..))))))))).-------........... ( -25.50) >DroPer_CAF1 95910 103 + 1 G-CGAAAUUGUCCUUUAAUUGGUUUUGCGCAUUAAUUUUGCACAAUUUAUUGCGCUUUGUGUGUUGUGCUGUGCAACUGCAAGU-----UGCAACUU---GCACAUUUGUG-U (-((((((((.........)))))))))...........((((((......((((.....))))(((((..((((((.....))-----))))....---))))).)))))-) ( -28.70) >consensus GCACAAAUUGUCCUUUAAUUGGUUUUGCGCAUUAAUUUUGCAGAAUUUAUUGCGCUUUGUGCUCUGCGCUUUGCA_CUGCAAGU_____UGCAAUUU____CGCAUUUGCAUU ...............((((..((((((((.........))))))))..)))).((...((((..(((.....)))..((((........)))).........))))..))... (-12.89 = -12.05 + -0.84)


| Location | 8,642,724 – 8,642,836 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.33 |
| Mean single sequence MFE | -24.17 |
| Consensus MFE | -10.02 |
| Energy contribution | -11.02 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8642724 112 - 22224390 AAUGCAAAUGCGAAUGAAAAUGCAGCCGGACUUGCAC-GGCAAAGCGGCGAGCACAAAGCGCAAUAAAUUUUGCAAAAUUAAUGCGCAAAACCAAUUAAAGGACAAUUUGUGC ..(((...((((........))))((((........)-)))...)))....((((((((((((...(((((....)))))..)))))....((.......))....))))))) ( -28.50) >DroPse_CAF1 108446 103 - 1 A-CACAAAUGUGC---AAGUUGCA-----ACUUGCAGAUGCACAGCACAACACACAAAGCGCAAUAAAUUGUGCAAAAUUAAUGCGCAAAACCAAUUAAAGGACAAUUUCG-C .-......(((((---(..((((.-----....)))).))))))..............(((.(((...(((((((.......)))))))..((.......))...))).))-) ( -22.40) >DroSim_CAF1 43665 112 - 1 AAUGCGAAUGCGAAUGAAAAUGCAGCAGGACUCGCAC-GGCAAAGCGCCGAGCACAAAGCGCAAUAAAUUCUGCAAAAUUAAUGCGCAAAACCAAUUAAAGGACAAUUUGUGC ..(((((.(((..((....))...)))....)))))(-(((.....)))).((((((((((((.(((.((.....)).))).)))))....((.......))....))))))) ( -32.50) >DroEre_CAF1 92020 88 - 1 AAUGCAAAUGC------------------------AG-UGCAAAACGCAGAGCACAAAGCGCAAUAAAUUCUGCAAAAUUAAUGCGCAAAACCAAUUAAAGGACAAUUUGUGG ..(((....))------------------------).-(((.....)))...(((((((((((.(((.((.....)).))).)))))....((.......))....)))))). ( -17.90) >DroYak_CAF1 82457 105 - 1 AAUGAAAAUGC-------ACCACAACCGAACUCGCAA-UGCAAAACGCAAAGCACAAAGCGCAAUAAAUUCUGCAAAAUUAAUGCGCAAAACCAAUUAAAGGACAAUUUGUGG ...........-------.((((((((......((..-(((.....)))..)).....(((((.(((.((.....)).))).))))).............)).....)))))) ( -19.50) >DroPer_CAF1 95910 103 - 1 A-CACAAAUGUGC---AAGUUGCA-----ACUUGCAGUUGCACAGCACAACACACAAAGCGCAAUAAAUUGUGCAAAAUUAAUGCGCAAAACCAAUUAAAGGACAAUUUCG-C .-......(((((---....((((-----((.....))))))..))))).........(((.(((...(((((((.......)))))))..((.......))...))).))-) ( -24.20) >consensus AAUGCAAAUGCG____AAAAUGCA_____ACUUGCAG_UGCAAAGCGCAAAGCACAAAGCGCAAUAAAUUCUGCAAAAUUAAUGCGCAAAACCAAUUAAAGGACAAUUUGUGC ....................................................(((((((((((.(((.((.....)).))).)))))....((.......))....)))))). (-10.02 = -11.02 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:00 2006