| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,616,139 – 8,616,236 |
| Length | 97 |
| Max. P | 0.995042 |

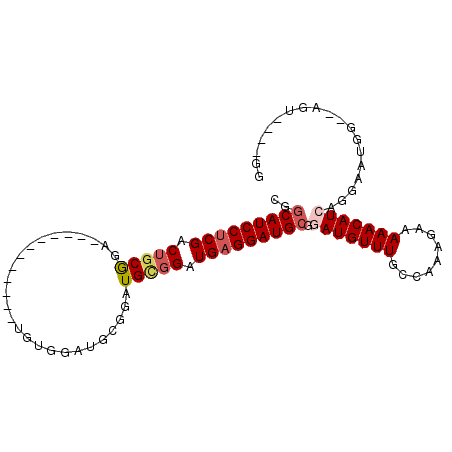
| Location | 8,616,139 – 8,616,236 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -19.59 |
| Consensus MFE | -11.89 |
| Energy contribution | -13.39 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8616139 97 + 22224390 CC----CCUCGCCAUUCCUCAUGUUUUUCUUUGGCAAACAUCCGCAUCCUCAUCCACAUCCGCAUCCACAUCCACAUUCACGUCCGCUGUCGAGGAUGCCG ..----....((((.................))))........((((((((....(((...((......................))))).)))))))).. ( -16.88) >DroSec_CAF1 53983 77 + 1 CC----ACU--CCAUGCCUGAUGUUUUUCUUUGGCAAACAUCCGCAUCCUCAUCCGCAUCC------ACA------------UCUGCAGUCGAGGAUGCCG ..----...--...((((.((......))...)))).......((((((((..(.(((...------...------------..))).)..)))))))).. ( -23.90) >DroEre_CAF1 66841 87 + 1 CCAGUCACU--CCAUUCCUGAUGUUUUUCUUUGGCAAACAUCCUCAUCCGCAUCCUCAUCCGCAUCCACA------------UCCGCAGUCGAGGAUGCCG .........--........(((((((.(.....).))))))).......((((((((..(.((.......------------...)).)..)))))))).. ( -20.70) >DroYak_CAF1 55021 83 + 1 CC----ACU--UCAUUCCUCAUGUUUUUCUUUGGCAAACAUCCGCAUCCUCAUCCGCAUCCGCAUCCACA------------UCCGCAGUCGAGGAUGCCG ..----...--.........((((((.(.....).))))))..((((((((..(.((.............------------...)).)..)))))))).. ( -16.89) >consensus CC____ACU__CCAUUCCUCAUGUUUUUCUUUGGCAAACAUCCGCAUCCUCAUCCGCAUCCGCAUCCACA____________UCCGCAGUCGAGGAUGCCG ...................(((((((.(.....).))))))).((((((((..(.((............................)).)..)))))))).. (-11.89 = -13.39 + 1.50)

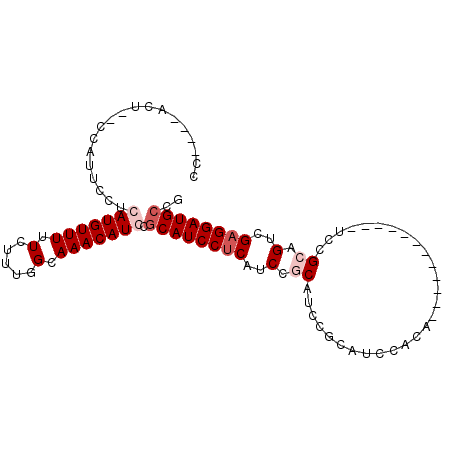

| Location | 8,616,139 – 8,616,236 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -18.68 |
| Energy contribution | -19.87 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8616139 97 - 22224390 CGGCAUCCUCGACAGCGGACGUGAAUGUGGAUGUGGAUGCGGAUGUGGAUGAGGAUGCGGAUGUUUGCCAAAGAAAAACAUGAGGAAUGGCGAGG----GG (.(((((((((.(.(((..(((................)))..))).).))))))))).).(.(((((((.................))))))).----). ( -28.42) >DroSec_CAF1 53983 77 - 1 CGGCAUCCUCGACUGCAGA------------UGU------GGAUGCGGAUGAGGAUGCGGAUGUUUGCCAAAGAAAAACAUCAGGCAUGG--AGU----GG ..(((((((((.(((((..------------...------...))))).))))))))).(((((((.........)))))))........--...----.. ( -29.60) >DroEre_CAF1 66841 87 - 1 CGGCAUCCUCGACUGCGGA------------UGUGGAUGCGGAUGAGGAUGCGGAUGAGGAUGUUUGCCAAAGAAAAACAUCAGGAAUGG--AGUGACUGG (.(((((((((.(((((..------------......))))).))))))))).).....(((((((.........)))))))........--......... ( -29.50) >DroYak_CAF1 55021 83 - 1 CGGCAUCCUCGACUGCGGA------------UGUGGAUGCGGAUGCGGAUGAGGAUGCGGAUGUUUGCCAAAGAAAAACAUGAGGAAUGA--AGU----GG (.(((((((((.(((((..------------(((....)))..))))).))))))))).)((((((.........)))))).........--...----.. ( -29.40) >consensus CGGCAUCCUCGACUGCGGA____________UGUGGAUGCGGAUGCGGAUGAGGAUGCGGAUGUUUGCCAAAGAAAAACAUCAGGAAUGG__AGU____GG ..(((((((((.(((((..........................))))).))))))))).(((((((.........)))))))................... (-18.68 = -19.87 + 1.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:56 2006