
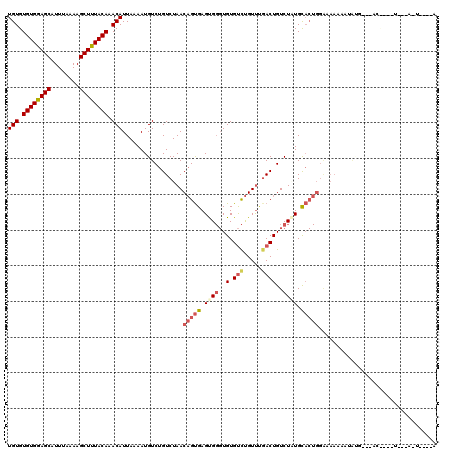
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,590,492 – 8,590,619 |
| Length | 127 |
| Max. P | 0.897532 |

| Location | 8,590,492 – 8,590,594 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.46 |
| Mean single sequence MFE | -26.64 |
| Consensus MFE | -17.60 |
| Energy contribution | -18.80 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8590492 102 - 22224390 UGUGUGUGAAGCAUUUAAAAGCUUUACAAACAUUAAAAUGUCUGUCUAACAGUGAGUGGGUGUGUCUGUUUGACUGUCUAUGCACUGGACAAAAAAAUGG--------------U----U (((.((((((((........)))))))).)))..........((((...(((((.((((..(.(((.....))))..)))).))))))))).........--------------.----. ( -28.90) >DroSec_CAF1 31052 116 - 1 UGUGUGUGAAGCAUUUAAAAGCUUUACAAACAUUAAAAUGUCUGUCUAACAGUGAGUGGGUGUGUCUGUUUGACUGUCUAUGCACUGGAAAAAAAGGGGGGAACCACAUAGUAAU----A .((.((((((((........)))))))).))((((..((((........(((((.((((..(.(((.....))))..)))).)))))..........((....))))))..))))----. ( -32.60) >DroEre_CAF1 41592 113 - 1 UGUGUGUGGAGCAUUUAAAAGCUUUACAAACAUUAAAAUGUCUGUCUAACAGUGAGUGGGUGUGUCUGUUUCACUGUCUCUGCACUGGGCAAAAUUACG--UACCGCAUUAAA-U----U ((((((((((((........))))))))..........((((((((..((((((((..((.....))..))))))))....).)).)))))........--...)))).....-.----. ( -25.30) >DroYak_CAF1 31232 117 - 1 UGUGUGUGGAGCAUUUAAAAGCUUUACAAACAUUAAAAUGUCUGUCUAACAGUGAGCGGGUGUGUCUGUUUGACUGUCUCUGCACUGGAAAAAACUAUG--UACUAUGUUCUA-UGGAUA (((.((((((((........)))))))).)))......(((((((..((((....(((((...(((.....)))...).))))(((((......))).)--)....))))..)-)))))) ( -26.50) >DroAna_CAF1 22442 82 - 1 UGUGUGUGGAGCAUUUAAAAGCUUUACAAACAUUAAAAUGUCUGUCUAACUGCGAGUG--UGUGCUCGUCCGACUGUCUGUGUC------------------------------------ (((.((((((((........)))))))).))).......(.(.(((.....((((((.--...))))))..))).).)......------------------------------------ ( -19.90) >consensus UGUGUGUGGAGCAUUUAAAAGCUUUACAAACAUUAAAAUGUCUGUCUAACAGUGAGUGGGUGUGUCUGUUUGACUGUCUAUGCACUGGAAAAAAAUAUG___AC____U___A_U____A (((.((((((((........)))))))).))).................(((((.((((..(.(((.....))))..)))).)))))................................. (-17.60 = -18.80 + 1.20)


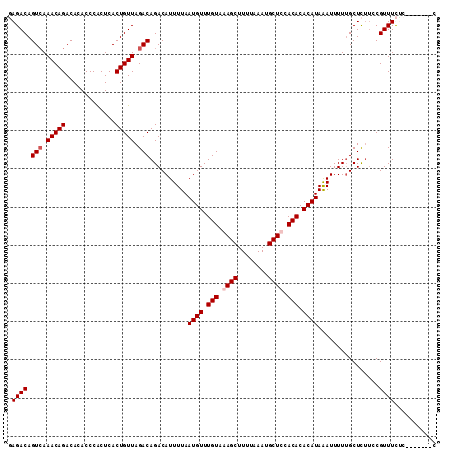
| Location | 8,590,514 – 8,590,619 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 90.32 |
| Mean single sequence MFE | -19.03 |
| Consensus MFE | -16.07 |
| Energy contribution | -16.82 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8590514 105 + 22224390 UAGACAGUCAAACAGACACACCCACUCACUGUUAGACAGACAUUUUAAUGUUUGUAAAGCUUUUAAAUGCUUCACACACAUAAAUUUUUGCUUCUCCGUUUCAC-------C .((((.(((.(((((.............))))).)))..........((((.(((.((((........)))).))).))))................))))...-------. ( -17.92) >DroSec_CAF1 31088 105 + 1 UAGACAGUCAAACAGACACACCCACUCACUGUUAGACAGACAUUUUAAUGUUUGUAAAGCUUUUAAAUGCUUCACACACAUAAAUUUUUGCUUUUCCGUUUCUC-------C .((((.(((.(((((.............))))).)))..........((((.(((.((((........)))).))).))))................))))...-------. ( -17.92) >DroEre_CAF1 41625 104 + 1 GAGACAGUGAAACAGACACACCCACUCACUGUUAGACAGACAUUUUAAUGUUUGUAAAGCUUUUAAAUGCUCCACACACAUAAAUUUUGGCUCUACCGUUUUUC-------- ..((((((((...............)))))))).((.((((......((((.(((..(((........)))..))).)))).......((.....)))))).))-------- ( -19.46) >DroYak_CAF1 31269 112 + 1 GAGACAGUCAAACAGACACACCCGCUCACUGUUAGACAGACAUUUUAAUGUUUGUAAAGCUUUUAAAUGCUCCACACACAUAGAUUUUUGCUCUUCCGUUUUCCGUUUCCCC (((((.(((.(((((.............))))).)))(((((..((.((((.(((..(((........)))..))).)))).))....)).)))..........)))))... ( -20.82) >consensus GAGACAGUCAAACAGACACACCCACUCACUGUUAGACAGACAUUUUAAUGUUUGUAAAGCUUUUAAAUGCUCCACACACAUAAAUUUUUGCUCUUCCGUUUCUC_______C .((((.(((.(((((.............))))).)))..........((((.(((.((((........)))).))).))))................))))........... (-16.07 = -16.82 + 0.75)


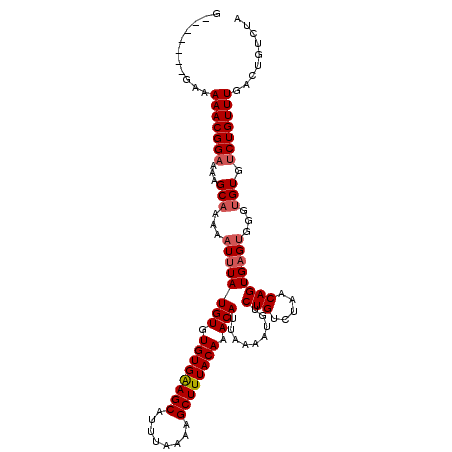
| Location | 8,590,514 – 8,590,619 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 90.32 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -21.02 |
| Energy contribution | -21.77 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8590514 105 - 22224390 G-------GUGAAACGGAGAAGCAAAAAUUUAUGUGUGUGAAGCAUUUAAAAGCUUUACAAACAUUAAAAUGUCUGUCUAACAGUGAGUGGGUGUGUCUGUUUGACUGUCUA (-------((.(((((((...(((...((((((((.((((((((........)))))))).))).........(((.....))))))))...))).))))))).)))..... ( -28.30) >DroSec_CAF1 31088 105 - 1 G-------GAGAAACGGAAAAGCAAAAAUUUAUGUGUGUGAAGCAUUUAAAAGCUUUACAAACAUUAAAAUGUCUGUCUAACAGUGAGUGGGUGUGUCUGUUUGACUGUCUA (-------(..(((((((...(((...((((((((.((((((((........)))))))).))).........(((.....))))))))...))).)))))))..))..... ( -25.30) >DroEre_CAF1 41625 104 - 1 --------GAAAAACGGUAGAGCCAAAAUUUAUGUGUGUGGAGCAUUUAAAAGCUUUACAAACAUUAAAAUGUCUGUCUAACAGUGAGUGGGUGUGUCUGUUUCACUGUCUC --------.......(..(((..((......((((.((((((((........)))))))).)))).....)))))..)..((((((((..((.....))..))))))))... ( -25.00) >DroYak_CAF1 31269 112 - 1 GGGGAAACGGAAAACGGAAGAGCAAAAAUCUAUGUGUGUGGAGCAUUUAAAAGCUUUACAAACAUUAAAAUGUCUGUCUAACAGUGAGCGGGUGUGUCUGUUUGACUGUCUC .((((.(((((...(....)...........((((.((((((((........)))))))).)))).......)))))....(((((((((((....))))))).)))))))) ( -27.90) >consensus G_______GAAAAACGGAAAAGCAAAAAUUUAUGUGUGUGAAGCAUUUAAAAGCUUUACAAACAUUAAAAUGUCUGUCUAACAGUGAGUGGGUGUGUCUGUUUGACUGUCUA ...........(((((((...(((...((((((((.((((((((........)))))))).))).........(((.....))))))))...))).)))))))......... (-21.02 = -21.77 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:48 2006