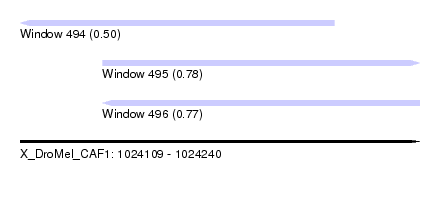
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,024,109 – 1,024,240 |
| Length | 131 |
| Max. P | 0.780926 |

| Location | 1,024,109 – 1,024,212 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.26 |
| Mean single sequence MFE | -26.57 |
| Consensus MFE | -14.87 |
| Energy contribution | -15.98 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.56 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1024109 103 - 22224390 UCUGUGG----GUGCGGGCAUUUUAAUUCAAUGCCCAAGUAAAUCAGCUUAAAGUGCAUGAAUAGUCUUUUCAUGGACGAUUUCAUCAAUUAUUCAAAGC---UG---G-ACCA------ ......(----((..(((((((.......))))))).......(((((((...((.((((((.......)))))).))(((...))).........))))---))---)-))).------ ( -30.30) >DroSec_CAF1 9623 103 - 1 UCUGUGG----GUGCGGGCAUUUUAAUUCAAUGCCCGAGUAAAUCAGCUUAAAGUGCAUGAAUAGUCUUUUCAUGGACGAUUUCAUCAAUUAUUCAAAGC---UG---G-ACCA------ ......(----((.((((((((.......))))))))......(((((((...((.((((((.......)))))).))(((...))).........))))---))---)-))).------ ( -32.00) >DroSim_CAF1 9186 103 - 1 UCUGUGG----GUGCGGGCAUUUUAAUUCAAUGCCCGAGUAAAUCAGCUUAAAGUGCAUGAAUAGUCUUUUCAUGGACGAUUUCAUCAAUUAUUCAAAGC---UG---G-ACCA------ ......(----((.((((((((.......))))))))......(((((((...((.((((((.......)))))).))(((...))).........))))---))---)-))).------ ( -32.00) >DroYak_CAF1 9977 107 - 1 UCUGUGGGUGGGUGCGGGCAUUUUAAUUCAAUGCCCGAGUAAAUCAGCUUAAAGUGCAUGAAUAGUCUUUUCAUGGACGAUUUCAUCAAUUAUUCAAAGC---UG---G-ACCA------ ..........(((.((((((((.......))))))))......(((((((...((.((((((.......)))))).))(((...))).........))))---))---)-))).------ ( -32.00) >DroAna_CAF1 14853 105 - 1 UCCUUG-----------GCAUUUUAAUUCGUUGG-CAAGUAAAUCAGCUUAAAGUGCAUGAAUGGCCACUUCAUGGACGAUUUCAUCAAUUAUUCAAAGC---CCAUAGCCCCAGCUCCG .....(-----------((..........(((((-........))))).....((.((((((.(....))))))).))....................))---)...(((....)))... ( -17.00) >DroPer_CAF1 128900 88 - 1 AGUGUAC----GUGUG-----------------G-UGUGUGCAUCAGCUUAAAGUGCAUGACUGGCCAAUUCAUGGACGAUUUCAUCAAUUAUUCAAAGCCUUUC---G-ACGG------ ......(----((.((-----------------(-((....)))))((((...((.(((((.(.....).))))).))(((...))).........)))).....---.-))).------ ( -16.10) >consensus UCUGUGG____GUGCGGGCAUUUUAAUUCAAUGCCCGAGUAAAUCAGCUUAAAGUGCAUGAAUAGUCUUUUCAUGGACGAUUUCAUCAAUUAUUCAAAGC___UG___G_ACCA______ ..............((((((((.......)))))))).........((((...((.((((((.......)))))).))(((...))).........)))).................... (-14.87 = -15.98 + 1.11)

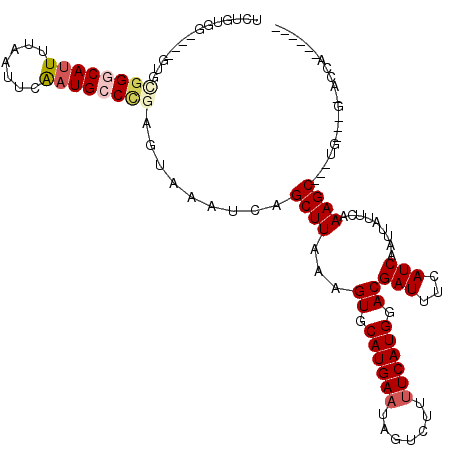
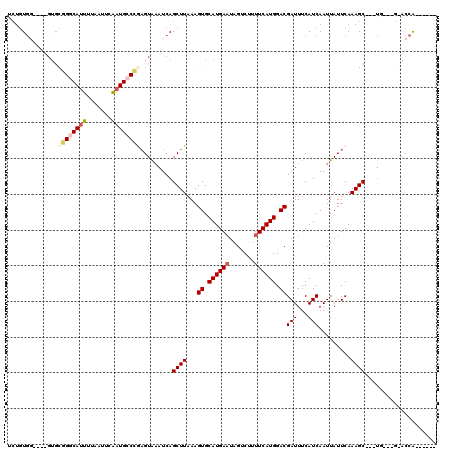
| Location | 1,024,136 – 1,024,240 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.97 |
| Mean single sequence MFE | -26.59 |
| Consensus MFE | -19.64 |
| Energy contribution | -20.41 |
| Covariance contribution | 0.77 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1024136 104 + 22224390 UCGUCCAUGAAAAGACUAUUCAUGCACUUUAAGCUGAUUUACUUGGGCAUUGAAUUAAAAUGCCCGCAC----CCACAGAGUGCAUUGACUGCCAAAGCAACACGACU------------ ..(((..............((((((((((((((........)))(((((((.......)))))))....----....)))))))).))).(((....)))....))).------------ ( -25.80) >DroSec_CAF1 9650 104 + 1 UCGUCCAUGAAAAGACUAUUCAUGCACUUUAAGCUGAUUUACUCGGGCAUUGAAUUAAAAUGCCCGCAC----CCACAGAGUGCAUUGACUGCCAAAGCAACACGACU------------ ..(((((((((.......))))))........(((.......(((((((((.......)))))))((((----.......))))...)).......))).....))).------------ ( -26.44) >DroSim_CAF1 9213 104 + 1 UCGUCCAUGAAAAGACUAUUCAUGCACUUUAAGCUGAUUUACUCGGGCAUUGAAUUAAAAUGCCCGCAC----CCACAGAGUGCAUUGACUGCCAAAGCAACACGACU------------ ..(((((((((.......))))))........(((.......(((((((((.......)))))))((((----.......))))...)).......))).....))).------------ ( -26.44) >DroYak_CAF1 10004 112 + 1 UCGUCCAUGAAAAGACUAUUCAUGCACUUUAAGCUGAUUUACUCGGGCAUUGAAUUAAAAUGCCCGCACCCACCCACAGAGUGCAUUGACUGGCAAAGCAACACGACUCGAC-------- ((((...((....(.(((.(((((((((((.............((((((((.......))))))))...........)))))))).))).))))....))..))))......-------- ( -27.65) >DroAna_CAF1 14890 108 + 1 UCGUCCAUGAAGUGGCCAUUCAUGCACUUUAAGCUGAUUUACUUG-CCAACGAAUUAAAAUGC-----------CAAGGAGUGCAUUGACUGGCUAGGAUAUAUAUGUAGAGGAUGUAGU .(((((......((((((.(((((((((((..((((((((.....-.....))))))....))-----------...)))))))).))).))))))...(((....)))..))))).... ( -26.60) >consensus UCGUCCAUGAAAAGACUAUUCAUGCACUUUAAGCUGAUUUACUCGGGCAUUGAAUUAAAAUGCCCGCAC____CCACAGAGUGCAUUGACUGCCAAAGCAACACGACU____________ ..(((.(((((.......)))))(((((((.............((((((((.......))))))))...........)))))))...))).............................. (-19.64 = -20.41 + 0.77)

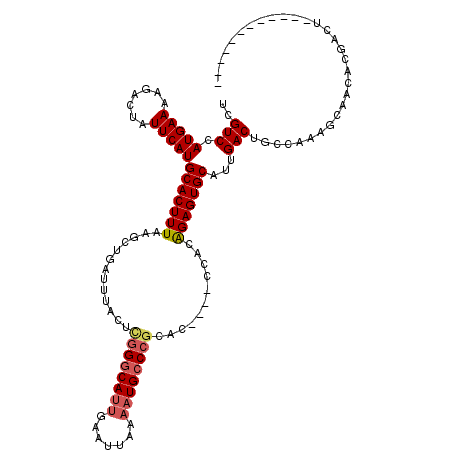
| Location | 1,024,136 – 1,024,240 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.97 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -20.93 |
| Energy contribution | -21.18 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1024136 104 - 22224390 ------------AGUCGUGUUGCUUUGGCAGUCAAUGCACUCUGUGG----GUGCGGGCAUUUUAAUUCAAUGCCCAAGUAAAUCAGCUUAAAGUGCAUGAAUAGUCUUUUCAUGGACGA ------------..((((((.((((((((.......(((((.....)----))))(((((((.......)))))))..........)).))))))))))))...((((......)))).. ( -34.50) >DroSec_CAF1 9650 104 - 1 ------------AGUCGUGUUGCUUUGGCAGUCAAUGCACUCUGUGG----GUGCGGGCAUUUUAAUUCAAUGCCCGAGUAAAUCAGCUUAAAGUGCAUGAAUAGUCUUUUCAUGGACGA ------------..((((((.((((((((.......(((((.....)----))))(((((((.......)))))))..........)).))))))))))))...((((......)))).. ( -34.50) >DroSim_CAF1 9213 104 - 1 ------------AGUCGUGUUGCUUUGGCAGUCAAUGCACUCUGUGG----GUGCGGGCAUUUUAAUUCAAUGCCCGAGUAAAUCAGCUUAAAGUGCAUGAAUAGUCUUUUCAUGGACGA ------------..((((((.((((((((.......(((((.....)----))))(((((((.......)))))))..........)).))))))))))))...((((......)))).. ( -34.50) >DroYak_CAF1 10004 112 - 1 --------GUCGAGUCGUGUUGCUUUGCCAGUCAAUGCACUCUGUGGGUGGGUGCGGGCAUUUUAAUUCAAUGCCCGAGUAAAUCAGCUUAAAGUGCAUGAAUAGUCUUUUCAUGGACGA --------..........((((.((((((.......((((((.......))))))(((((((.......)))))))).))))).)))).....((.((((((.......)))))).)).. ( -35.80) >DroAna_CAF1 14890 108 - 1 ACUACAUCCUCUACAUAUAUAUCCUAGCCAGUCAAUGCACUCCUUG-----------GCAUUUUAAUUCGUUGG-CAAGUAAAUCAGCUUAAAGUGCAUGAAUGGCCACUUCAUGGACGA .....................(((..((((.(((.((((((.((((-----------.((...........)).-))))(((......))).))))))))).))))........)))... ( -19.80) >consensus ____________AGUCGUGUUGCUUUGGCAGUCAAUGCACUCUGUGG____GUGCGGGCAUUUUAAUUCAAUGCCCGAGUAAAUCAGCUUAAAGUGCAUGAAUAGUCUUUUCAUGGACGA ..............................(((...(((((..............(((((((.......)))))))((((......))))..)))))(((((.......))))).))).. (-20.93 = -21.18 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:48 2006