
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,555,075 – 8,555,233 |
| Length | 158 |
| Max. P | 0.792034 |

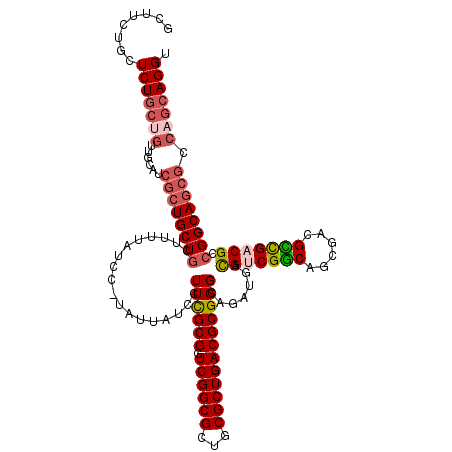
| Location | 8,555,075 – 8,555,195 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -50.59 |
| Consensus MFE | -37.62 |
| Energy contribution | -38.94 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8555075 120 + 22224390 GCUUUUGCUCUGCUGUUGGAUCGCUGCUGCUAUUAUCCCUAUUAUCCUUCGCGGUCGGCGCUGCGCUGACGCGGGCAGAUGACGUCGGCAGCGACGCCGACGCCGGCAGCGCCAGCAGGU ((....))....(((((((..((((((((............(((((.((((((.((((((...))))))))))))..)))))(((((((......))))))).))))))))))))))).. ( -56.20) >DroSec_CAF1 15832 120 + 1 GCUUCUGCUCUGCCGUUGCAUCGAUGCUGCUAUUAUCCUUAUUAUCCUUCGCGGUCGGCGCAGCGCUGACGCGGGCAGAUGACGUCGGCAGCGACGCCGACGCCGGCAGCGCCAGCAGGU ...(((((((((((((.(((....))).))....................(((.((((((..(((((((((((......)).))))))).))..))))))))).)))))....)))))). ( -50.80) >DroSim_CAF1 23042 120 + 1 GCUUCUGCUCUGCCGUUUCAUCGCUGCUGCUUUUAUCCUUAUUAUCCUUCGCGGUCGGCGCUGCGCUGACGCGGGCAGAUGACGUCGGCAGCGACGCCGACGCCGGCAGCGCCAGCAGGU ...((((((((((((..(((((((((((((....................)))).))))(((.(((....)))))).)))))(((((((......))))))).))))))....)))))). ( -51.95) >DroEre_CAF1 16717 108 + 1 GCUUCUGCUCUGCUGCUGCUUCGCUGCUUCUU------------UGCUUUGCGGUCGGCGCAGCGCUGACGCGGGCAGAUGAUGCCGGCAGCGACGUCGACGCCGGCAGCGCCAGCAGGU ...((((((.(((((((((..(((.((.....------------.))...)))(((((((...((((...((.((((.....)))).)))))).)))))))).))))))))..)))))). ( -52.60) >DroYak_CAF1 17978 100 + 1 AUUUUAGAUCUUAUC--------CUGCUGCUU------------UGCUUCGCGGUCGGCGCUGCGCUGACGCGGGCAGAUGACGGCGACAGUGACGUUGACGCCGGCAGCGCCAGUAGGU .........(((((.--------.((((((((------------((((.((((.((((((...))))))))))))))))...(((((.(((.....))).)))))))))))...))))). ( -41.40) >consensus GCUUCUGCUCUGCUGUUGCAUCGCUGCUGCUUUUAUCC_UAUUAUCCUUCGCGGUCGGCGCUGCGCUGACGCGGGCAGAUGACGUCGGCAGCGACGCCGACGCCGGCAGCGCCAGCAGGU ........(((((((......((((((((..................((((((.((((((...)))))))))))).......(((((((......))))))).)))))))).))))))). (-37.62 = -38.94 + 1.32)

| Location | 8,555,075 – 8,555,195 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -47.96 |
| Consensus MFE | -36.64 |
| Energy contribution | -37.64 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8555075 120 - 22224390 ACCUGCUGGCGCUGCCGGCGUCGGCGUCGCUGCCGACGUCAUCUGCCCGCGUCAGCGCAGCGCCGACCGCGAAGGAUAAUAGGGAUAAUAGCAGCAGCGAUCCAACAGCAGAGCAAAAGC ..(((((((((((((.((((((((((....))))))))))........((....))))))))))..((.....)).......((((....((....)).))))...)))))......... ( -52.60) >DroSec_CAF1 15832 120 - 1 ACCUGCUGGCGCUGCCGGCGUCGGCGUCGCUGCCGACGUCAUCUGCCCGCGUCAGCGCUGCGCCGACCGCGAAGGAUAAUAAGGAUAAUAGCAGCAUCGAUGCAACGGCAGAGCAGAAGC ..((((.(((...)))((((((((((....)))))))))).((((((.(((((...(((((.((.....(....).......))......)))))...)))))...)))))))))).... ( -52.90) >DroSim_CAF1 23042 120 - 1 ACCUGCUGGCGCUGCCGGCGUCGGCGUCGCUGCCGACGUCAUCUGCCCGCGUCAGCGCAGCGCCGACCGCGAAGGAUAAUAAGGAUAAAAGCAGCAGCGAUGAAACGGCAGAGCAGAAGC ..((((((.((((((.((((((((((....))))))))))((((...((((((.(((...))).)).))))..))))................))))))......))))))......... ( -50.90) >DroEre_CAF1 16717 108 - 1 ACCUGCUGGCGCUGCCGGCGUCGACGUCGCUGCCGGCAUCAUCUGCCCGCGUCAGCGCUGCGCCGACCGCAAAGCA------------AAGAAGCAGCGAAGCAGCAGCAGAGCAGAAGC ..(((((.(((((((((((((.(.(((.(.(((.((((.....)))).))).).)))).))))))..(((...((.------------.....)).)))..))))).))..))))).... ( -47.40) >DroYak_CAF1 17978 100 - 1 ACCUACUGGCGCUGCCGGCGUCAACGUCACUGUCGCCGUCAUCUGCCCGCGUCAGCGCAGCGCCGACCGCGAAGCA------------AAGCAGCAG--------GAUAAGAUCUAAAAU .(((.(((((((((((((((.((.......)).)))))..........((....))))))))))....((......------------..)))).))--------).............. ( -36.00) >consensus ACCUGCUGGCGCUGCCGGCGUCGGCGUCGCUGCCGACGUCAUCUGCCCGCGUCAGCGCAGCGCCGACCGCGAAGGAUAAUA_GGAUAAAAGCAGCAGCGAUGCAACAGCAGAGCAGAAGC ..(((((((((((((.((((((((((....))))))))))........((....))))))))))....((.......................))...........)))))......... (-36.64 = -37.64 + 1.00)



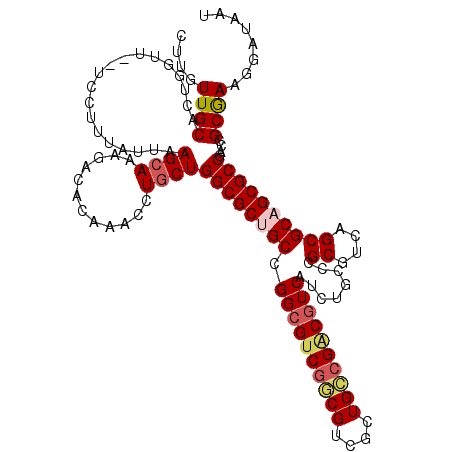
| Location | 8,555,115 – 8,555,233 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.67 |
| Mean single sequence MFE | -44.00 |
| Consensus MFE | -36.20 |
| Energy contribution | -37.08 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8555115 118 - 22224390 CUUGUUGCACUGAUU--UCCUUUAUGAAGCAAAGACACAAACCUGCUGGCGCUGCCGGCGUCGGCGUCGCUGCCGACGUCAUCUGCCCGCGUCAGCGCAGCGCCGACCGCGAAGGAUAAU ...............--(((((..((.((((............))))((((((((.((((((((((....))))))))))........((....))))))))))...))..))))).... ( -47.90) >DroSec_CAF1 15872 120 - 1 CUUGUUGCACUAGUUGCUCCUUUAUUAAGCAAAGACACAAACCUGCUGGCGCUGCCGGCGUCGGCGUCGCUGCCGACGUCAUCUGCCCGCGUCAGCGCUGCGCCGACCGCGAAGGAUAAU ......(((.....)))((((((....(((..((........))((((((((.((.((((((((((....))))))))))....))..)))))))))))(((.....))))))))).... ( -45.10) >DroSim_CAF1 23082 120 - 1 CUUGUUGCACUAGUUGCUCCUUUAUUAAGCAAAGACACAAACCUGCUGGCGCUGCCGGCGUCGGCGUCGCUGCCGACGUCAUCUGCCCGCGUCAGCGCAGCGCCGACCGCGAAGGAUAAU ......(((.....)))((((((....((((............))))((((((((.((((((((((....))))))))))........((....))))))))))......)))))).... ( -47.80) >DroEre_CAF1 16749 114 - 1 CUUGUUGCACUGGUU--UCCUUUAUUAAGCAGAGACACAAACCUGCUGGCGCUGCCGGCGUCGACGUCGCUGCCGGCAUCAUCUGCCCGCGUCAGCGCUGCGCCGACCGCAAAGCA---- ....((((...((((--..........(((((..........)))))(((((((((((((.((....)).))))))))..........(((....))).)))))))))))))....---- ( -43.00) >DroYak_CAF1 18002 114 - 1 CUUGUUGCACUGGUU--UCCUUAAUUAAGCAAAGACACGAACCUACUGGCGCUGCCGGCGUCAACGUCACUGUCGCCGUCAUCUGCCCGCGUCAGCGCAGCGCCGACCGCGAAGCA---- ....((((...((((--(.(((.........)))....)))))...((((((((((((((.((.......)).)))))..........((....)))))))))))...))))....---- ( -36.20) >consensus CUUGUUGCACUGGUU__UCCUUUAUUAAGCAAAGACACAAACCUGCUGGCGCUGCCGGCGUCGGCGUCGCUGCCGACGUCAUCUGCCCGCGUCAGCGCAGCGCCGACCGCGAAGGAUAAU ....((((...................((((............))))((((((((.((((((((((....))))))))))........((....))))))))))....))))........ (-36.20 = -37.08 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:39 2006