
| Sequence ID | X_DroMel_CAF1 |
|---|---|
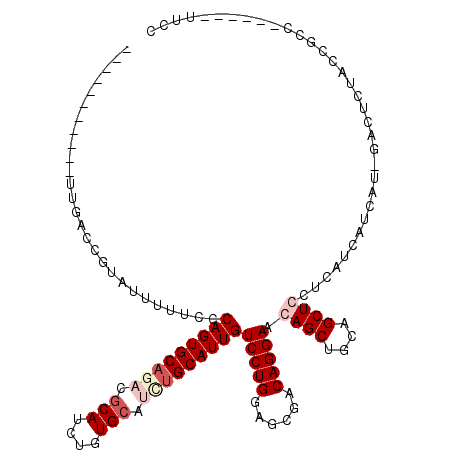
| Location | 8,552,544 – 8,552,658 |
| Length | 114 |
| Max. P | 0.900436 |

| Location | 8,552,544 – 8,552,658 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.23 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -20.14 |
| Energy contribution | -22.07 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
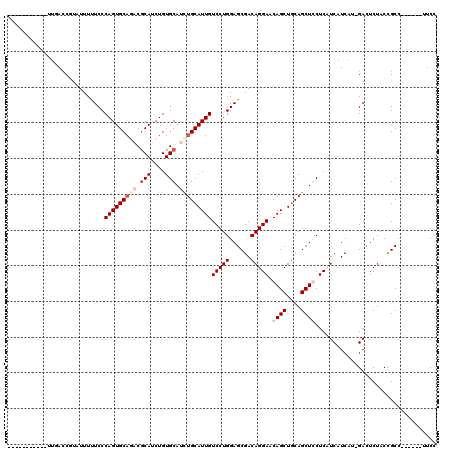
>X_DroMel_CAF1 8552544 114 + 22224390 CAGUAAUUCUUUUGACCUUAUUUUUCCCAGUGCAGACGCAUCUGUGCAUCUGCAUUGUCCUGGAGCGACAGGAACAGCUGCAGCUCCUCAUCAUCAUAGAGUCUACCGCC------UUCC ..((((((((..(((............(((((((((.(((....))).)))))))))....(((((..(((......)))..)))))......))).))))).)))....------.... ( -35.10) >DroSec_CAF1 13256 99 + 1 --------------ACCGUUUUUUUCCCAGUGCAGACGCAUCUGUGCAUUUGCAUUGUCCUGGAGCGACAGGAACAGCUGCAGCUCCUCAUCAUCAU-GACUCUACUGCC------UUCG --------------.............(((((((((.(((....))).)))))))))....(((((..(((......)))..)))))((((....))-))..........------.... ( -27.10) >DroEre_CAF1 14021 102 + 1 -----------UUGACCGUAUUUUUCCCAGUGCCUCCACAUCUGUGCAUCUGCAUUGUCCUGGAGCGACAGGACCAGCUACAGCUGCUCCUCAUCAU-GACUCUACCGCC------UUCC -----------.................((.(((((((...(.((((....)))).)...))))).((.((((.((((....)))).))))..))..-.........)))------)... ( -23.10) >DroYak_CAF1 15144 107 + 1 -----------UUCA-CUAAUUUAUCCCAGUGCAUCCGCAUCUGUGCAUCUGCAUUGUCCUGGAGCGACAGGACCAGCUGCAGCUGCUCCUCAUCAU-GACUCUACCGCCCACAUCCUCC -----------....-...........(((((((...(((....)))...)))))))...(((.(((...(((.((((....)))).)))((.....-))......))))))........ ( -26.40) >consensus ___________UUGACCGUAUUUUUCCCAGUGCAGACGCAUCUGUGCAUCUGCAUUGUCCUGGAGCGACAGGAACAGCUGCAGCUCCUCAUCAUCAU_GACUCUACCGCC______UUCC ...........................(((((((((.(((....))).)))))))))(((((......))))).((((....)))).................................. (-20.14 = -22.07 + 1.94)

| Location | 8,552,544 – 8,552,658 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.23 |
| Mean single sequence MFE | -33.85 |
| Consensus MFE | -20.58 |
| Energy contribution | -20.95 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
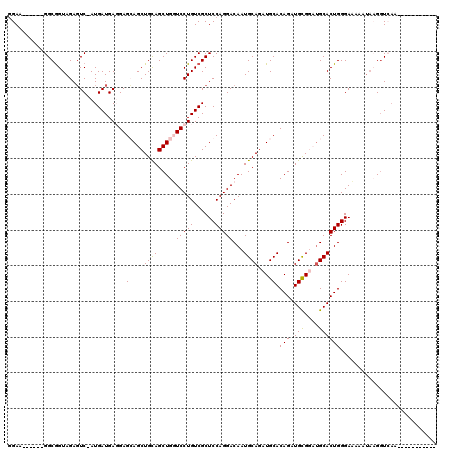
>X_DroMel_CAF1 8552544 114 - 22224390 GGAA------GGCGGUAGACUCUAUGAUGAUGAGGAGCUGCAGCUGUUCCUGUCGCUCCAGGACAAUGCAGAUGCACAGAUGCGUCUGCACUGGGAAAAAUAAGGUCAAAAGAAUUACUG ....------..((((((..(((.((((.....(((((.((((......)))).)))))....((.((((((((((....)))))))))).))...........))))..))).)))))) ( -37.70) >DroSec_CAF1 13256 99 - 1 CGAA------GGCAGUAGAGUC-AUGAUGAUGAGGAGCUGCAGCUGUUCCUGUCGCUCCAGGACAAUGCAAAUGCACAGAUGCGUCUGCACUGGGAAAAAAACGGU-------------- ....------.(((((....((-((....))))...))))).((((((((((......)))))((.((((.(((((....))))).)))).))........)))))-------------- ( -28.30) >DroEre_CAF1 14021 102 - 1 GGAA------GGCGGUAGAGUC-AUGAUGAGGAGCAGCUGUAGCUGGUCCUGUCGCUCCAGGACAAUGCAGAUGCACAGAUGUGGAGGCACUGGGAAAAAUACGGUCAA----------- ....------(((.(((...((-..(((.((((.((((....)))).)))))))((((((.(.(..(((....)))..).).)))).)).....))....))).)))..----------- ( -34.30) >DroYak_CAF1 15144 107 - 1 GGAGGAUGUGGGCGGUAGAGUC-AUGAUGAGGAGCAGCUGCAGCUGGUCCUGUCGCUCCAGGACAAUGCAGAUGCACAGAUGCGGAUGCACUGGGAUAAAUUAG-UGAA----------- (((((((...(((......)))-......((((.((((....)))).))))))).))))....((.((((..((((....))))..)))).))...........-....----------- ( -35.10) >consensus GGAA______GGCGGUAGAGUC_AUGAUGAGGAGCAGCUGCAGCUGGUCCUGUCGCUCCAGGACAAUGCAGAUGCACAGAUGCGGAUGCACUGGGAAAAAUAAGGUCAA___________ .................................(((.(((((..((.(((((......))))))).))))).))).(((.(((....))))))........................... (-20.58 = -20.95 + 0.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:32 2006