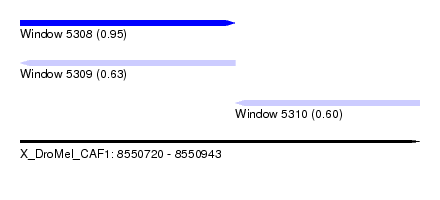
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,550,720 – 8,550,943 |
| Length | 223 |
| Max. P | 0.954147 |

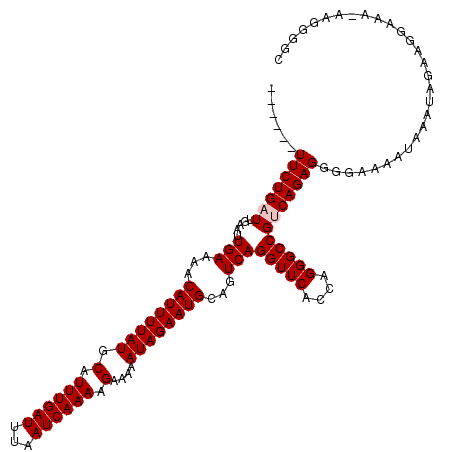
| Location | 8,550,720 – 8,550,840 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.26 |
| Mean single sequence MFE | -22.63 |
| Consensus MFE | -20.32 |
| Energy contribution | -20.72 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8550720 120 + 22224390 UUUUUGUUCUGAUUGAAUUGAAAACAUUUUAUGCAUUUGAUUUAAUCAAAAGAAAAAUAGAAUGCAGUCAGGUUCACCAGGGCCGUCAGAGGGGAAAAUAAAUAGAAGGAAAAAAGGGGC (((((.(((((((.....(((...((((((((.(.((((((...)))))).)....))))))))...)))(((((....)))))))))))).)))))....................... ( -26.30) >DroSec_CAF1 11530 113 + 1 ------UUCUGAUUGAAUUGAAAACAUUUUAUGCAUUUGAUUUAAUCAAAAGAAAAAUAGAAUGCAGUCAGGUUCACCAGGGCCGACAGAGGGGAAAAUAAAUAGAAGGAAA-AAGGGGC ------(((((.......(((...((((((((.(.((((((...)))))).)....))))))))...)))(((((....)))))..))))).....................-....... ( -20.20) >DroSim_CAF1 17740 113 + 1 ------UUCUGAUUGAAUUGAAAACAUUUUAUGCAUUUGAUUUAAUCAAAAGAAAAAUAGAAUGCAGUCAGGUUCACCAGGGCCGACAGAGGGGAAAAUAAAUAGAAGGAAA-AAGGGGC ------(((((.......(((...((((((((.(.((((((...)))))).)....))))))))...)))(((((....)))))..))))).....................-....... ( -20.20) >DroEre_CAF1 12134 109 + 1 ------UUCUGAUUGAAUUGAAAACAUUUUAUGCAUUUGAUUUAAUCAAAAGAAAAAUAGAAUGCAGUCAGGUUCCCCAGGGCCGUCAGAGGGGAAAA---AGA-GCGGGAA-AGGGGUG ------.((((((((.........((((((((.(.((((((...)))))).)....))))))))))))))))..((((....((((............---...-))))...-.)))).. ( -24.16) >DroYak_CAF1 13260 110 + 1 ------UUCUGAUUGAAUUGAAAACAUUUUAUGCAUUUGAUUUAAUCAAAAGAAAAAUAGAAUGCAGUCAGGUUCACCAGGGCCGUCAGAGGGAAAAA---AGAAGAGGAAA-AAGGGGG ------(((((((.....(((...((((((((.(.((((((...)))))).)....))))))))...)))(((((....)))))))))))).......---...........-....... ( -22.30) >consensus ______UUCUGAUUGAAUUGAAAACAUUUUAUGCAUUUGAUUUAAUCAAAAGAAAAAUAGAAUGCAGUCAGGUUCACCAGGGCCGUCAGAGGGGAAAAUAAAUAGAAGGAAA_AAGGGGC ......(((((((.....(((...((((((((.(.((((((...)))))).)....))))))))...)))(((((....))))))))))))............................. (-20.32 = -20.72 + 0.40)

| Location | 8,550,720 – 8,550,840 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.26 |
| Mean single sequence MFE | -15.74 |
| Consensus MFE | -13.02 |
| Energy contribution | -13.42 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8550720 120 - 22224390 GCCCCUUUUUUCCUUCUAUUUAUUUUCCCCUCUGACGGCCCUGGUGAACCUGACUGCAUUCUAUUUUUCUUUUGAUUAAAUCAAAUGCAUAAAAUGUUUUCAAUUCAAUCAGAACAAAAA ..............................(((((.......((....))(((..(((((.(((......((((((...))))))...))).)))))..)))......)))))....... ( -14.60) >DroSec_CAF1 11530 113 - 1 GCCCCUU-UUUCCUUCUAUUUAUUUUCCCCUCUGUCGGCCCUGGUGAACCUGACUGCAUUCUAUUUUUCUUUUGAUUAAAUCAAAUGCAUAAAAUGUUUUCAAUUCAAUCAGAA------ (((....-............................))).((((((((..(((..(((((.(((......((((((...))))))...))).)))))..))).))).)))))..------ ( -14.35) >DroSim_CAF1 17740 113 - 1 GCCCCUU-UUUCCUUCUAUUUAUUUUCCCCUCUGUCGGCCCUGGUGAACCUGACUGCAUUCUAUUUUUCUUUUGAUUAAAUCAAAUGCAUAAAAUGUUUUCAAUUCAAUCAGAA------ (((....-............................))).((((((((..(((..(((((.(((......((((((...))))))...))).)))))..))).))).)))))..------ ( -14.35) >DroEre_CAF1 12134 109 - 1 CACCCCU-UUCCCGC-UCU---UUUUCCCCUCUGACGGCCCUGGGGAACCUGACUGCAUUCUAUUUUUCUUUUGAUUAAAUCAAAUGCAUAAAAUGUUUUCAAUUCAAUCAGAA------ ..((((.-...(((.-((.---...........)))))....))))...((((.((((((..((((.((....))..))))..)))))).....((....))......))))..------ ( -20.80) >DroYak_CAF1 13260 110 - 1 CCCCCUU-UUUCCUCUUCU---UUUUUCCCUCUGACGGCCCUGGUGAACCUGACUGCAUUCUAUUUUUCUUUUGAUUAAAUCAAAUGCAUAAAAUGUUUUCAAUUCAAUCAGAA------ .......-...........---........(((((.......((....))(((..(((((.(((......((((((...))))))...))).)))))..)))......))))).------ ( -14.60) >consensus GCCCCUU_UUUCCUUCUAUUUAUUUUCCCCUCUGACGGCCCUGGUGAACCUGACUGCAUUCUAUUUUUCUUUUGAUUAAAUCAAAUGCAUAAAAUGUUUUCAAUUCAAUCAGAA______ ..............................(((((.......((....))(((..(((((.(((......((((((...))))))...))).)))))..)))......)))))....... (-13.02 = -13.42 + 0.40)

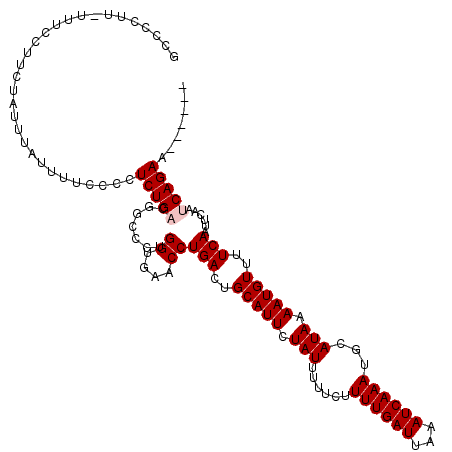
| Location | 8,550,840 – 8,550,943 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.79 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -19.56 |
| Energy contribution | -19.96 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8550840 103 - 22224390 GGGGACCGUCAAACAAUGCAAAUUGCAUGAAUGAAUGCCAUUUUUCAUUUCAUUUCAAUGCCCAGCACC-----------------CCACCACCCCCUUGACCCCGUUGACCCCCGUGCA ((((...(((((......(((((((.(((((((((........)))).)))))..))))((...))...-----------------...........)))......)))))))))..... ( -19.30) >DroSec_CAF1 11643 94 - 1 GGGGACCGUCAAACAAUGCAAAUUGCAUGAAUGAAUGCCAUUUUGCAUUUCAUUUCAAUGCCCAC-------------------------CGCCCCC-UGACCCCGUUGACCCCCGUGCA ((((...(((((..((((((((..((((......))))...)))))))).....(((..((....-------------------------.))....-))).....)))))))))..... ( -23.20) >DroSim_CAF1 17853 101 - 1 NGGGACCGUCAAACAAUGCAAAUUGCAUGAAUGAAUGCCAUUUUGCAUUUCAUUUCAAUGCCCACCGC------------------CCACCGCCCCC-UGACCCCGUUGACCCCCGUGCA .(((...(((((..((((((((..((((......))))...)))))))).................((------------------.....))....-........))))).)))..... ( -20.50) >DroEre_CAF1 12243 94 - 1 GGGGACCGUCAAACAAUGCAAAUUGCAUGAAUGAAUGCCAUUUUGCAUUUCAUUUCAAUGCCCACCA-------------------------CCCCC-UGACCCCUUUGACCCCCGUGCG ((((...((((((..((((.....))))((((((((((......))).)))))))............-------------------------.....-.......))))))))))..... ( -22.40) >DroYak_CAF1 13370 119 - 1 GGGGACCGUCAAACAAUGCAAAUUGCAUGAAUGAAUGCCAUUUUGCAUUUCAUUUCAAUGCCCACCAACCCACCAAAGCAACCAACCCACCGCCCCC-UGACCCCUUUGACCCCCGUGCA ((((...((((((.((((((((..((((......))))...)))))))).........(((................))).................-.......))))))))))..... ( -24.09) >consensus GGGGACCGUCAAACAAUGCAAAUUGCAUGAAUGAAUGCCAUUUUGCAUUUCAUUUCAAUGCCCACCA___________________CCACCGCCCCC_UGACCCCGUUGACCCCCGUGCA ((((...(((((...((((.....))))((((((((((......))).)))))))...................................................)))))))))..... (-19.56 = -19.96 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:30 2006