| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,548,064 – 8,548,259 |
| Length | 195 |
| Max. P | 0.995992 |

| Location | 8,548,064 – 8,548,184 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.02 |
| Mean single sequence MFE | -34.66 |
| Consensus MFE | -27.34 |
| Energy contribution | -27.78 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.995009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8548064 120 + 22224390 GAAUCGGAUGUUGCAUCAAUUAUAUUGAUUGCAUUUAGCAUUGCAUUGCAGGUGCGUUGGAAAAGUGCAAUAAUGCAAAUGCAUUUUGCUUGCAAUUUUGAUUUUGAUUGGGCUGCGAUA ........((((((((((((((.((((((((((...((((.(((((((((..((((((....)).))))....))).))))))...)))))))))))..)))..)))))))..))))))) ( -35.90) >DroSec_CAF1 9004 119 + 1 GAAUCGGAUGUUGCAUCAAUUAUAUUGAUUGCAUUU-GCAUUGCAUUGCUGGUGCGUUGGAAAAGUGCAAUAAUGCAAAUGCAUUUUGCUUGCAAUUUUGAUUUUGAUUGGGCUGUGAUA ((((((((.(((((((((((...))))).(((((((-((((((((((.(..(....)..)...))))))...))))))))))).......))))))))))))))................ ( -33.60) >DroSim_CAF1 11877 119 + 1 GAAUCGGAUGUUGCAUCAAUUAUAUUGAUUGCAUUU-GCAUUGCAUUGCUGGUGCGUUGGAAAAGUGCAAUAAUGCAAAUGCAUUUUGCUUGCAAUUUUGAUUUUGAUUGGGCUGUGAUA ((((((((.(((((((((((...))))).(((((((-((((((((((.(..(....)..)...))))))...))))))))))).......))))))))))))))................ ( -33.60) >DroEre_CAF1 9580 108 + 1 GAAUCGGAUGUUGCAUCAAUUAUAUUGAUUGCAUUU-GCA-----UUGCAGGUGCGUUGGAAAAGUGCAAUAAUGCAAAUGCAUUUUGCUUGCAAUUUU------GAUUGAGUUGCGAUA ........((((((((((((((....(((((((...-(((-----.((((..(((((((...........)))))))..))))...))).))))))).)------))))))..))))))) ( -35.60) >DroYak_CAF1 10670 114 + 1 GAAUCGGAUGUUGCAUCAAUUAUAUUGAUUGCAUUU-GCA-----UUGCAGGUGCGUUGGAAAAGUGCAAUAAUGCAAAUGCAUUUUGCUUGCAAUUUUGAUUUUGAUUGCGUUGCGAUA ((((((((.(((((((((((...))))).(((((((-(((-----(((...(..(.((....)))..)..))))))))))))).......))))))))))))))..(((((...))))). ( -34.60) >consensus GAAUCGGAUGUUGCAUCAAUUAUAUUGAUUGCAUUU_GCAUUGCAUUGCAGGUGCGUUGGAAAAGUGCAAUAAUGCAAAUGCAUUUUGCUUGCAAUUUUGAUUUUGAUUGGGCUGCGAUA ........((((((((((((((....(((((((....(((......((((..(((((((...........)))))))..))))...))).))))))).......)))))))..))))))) (-27.34 = -27.78 + 0.44)


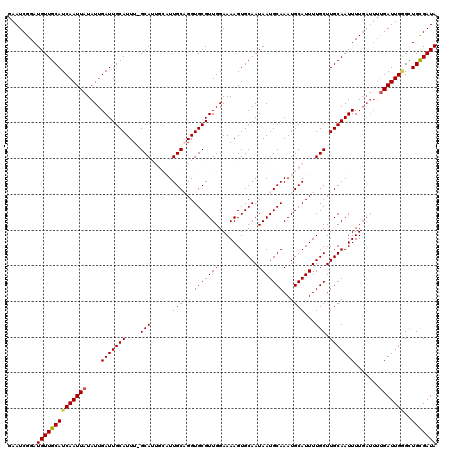
| Location | 8,548,064 – 8,548,184 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.02 |
| Mean single sequence MFE | -24.77 |
| Consensus MFE | -19.36 |
| Energy contribution | -19.36 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.10 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.995992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8548064 120 - 22224390 UAUCGCAGCCCAAUCAAAAUCAAAAUUGCAAGCAAAAUGCAUUUGCAUUAUUGCACUUUUCCAACGCACCUGCAAUGCAAUGCUAAAUGCAAUCAAUAUAAUUGAUGCAACAUCCGAUUC .((((...................((((((.(((((((((....))))).))))...........((....))..))))))......((((.(((((...))))))))).....)))).. ( -24.40) >DroSec_CAF1 9004 119 - 1 UAUCACAGCCCAAUCAAAAUCAAAAUUGCAAGCAAAAUGCAUUUGCAUUAUUGCACUUUUCCAACGCACCAGCAAUGCAAUGC-AAAUGCAAUCAAUAUAAUUGAUGCAACAUCCGAUUC .................((((....((((........(((((((((((((((((.................))))))..))))-)))))))((((((...)))))))))).....)))). ( -25.03) >DroSim_CAF1 11877 119 - 1 UAUCACAGCCCAAUCAAAAUCAAAAUUGCAAGCAAAAUGCAUUUGCAUUAUUGCACUUUUCCAACGCACCAGCAAUGCAAUGC-AAAUGCAAUCAAUAUAAUUGAUGCAACAUCCGAUUC .................((((....((((........(((((((((((((((((.................))))))..))))-)))))))((((((...)))))))))).....)))). ( -25.03) >DroEre_CAF1 9580 108 - 1 UAUCGCAACUCAAUC------AAAAUUGCAAGCAAAAUGCAUUUGCAUUAUUGCACUUUUCCAACGCACCUGCAA-----UGC-AAAUGCAAUCAAUAUAAUUGAUGCAACAUCCGAUUC .(((((((.......------....))))..((....((((((((((((.(((........))).((....))))-----)))-)))))))((((((...)))))))).......))).. ( -23.50) >DroYak_CAF1 10670 114 - 1 UAUCGCAACGCAAUCAAAAUCAAAAUUGCAAGCAAAAUGCAUUUGCAUUAUUGCACUUUUCCAACGCACCUGCAA-----UGC-AAAUGCAAUCAAUAUAAUUGAUGCAACAUCCGAUUC .((((....(((((..........)))))..((....((((((((((((.(((........))).((....))))-----)))-)))))))((((((...))))))))......)))).. ( -25.90) >consensus UAUCGCAGCCCAAUCAAAAUCAAAAUUGCAAGCAAAAUGCAUUUGCAUUAUUGCACUUUUCCAACGCACCUGCAAUGCAAUGC_AAAUGCAAUCAAUAUAAUUGAUGCAACAUCCGAUUC ...........((((............(((.(((((((((....))))).))))...........((....)).......)))....((((.(((((...)))))))))......)))). (-19.36 = -19.36 + -0.00)

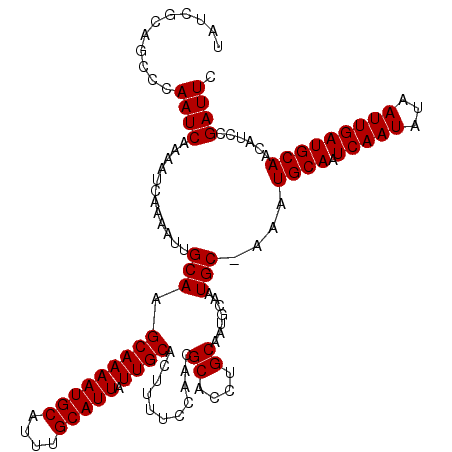

| Location | 8,548,104 – 8,548,224 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.58 |
| Mean single sequence MFE | -30.24 |
| Consensus MFE | -19.36 |
| Energy contribution | -19.52 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8548104 120 + 22224390 UUGCAUUGCAGGUGCGUUGGAAAAGUGCAAUAAUGCAAAUGCAUUUUGCUUGCAAUUUUGAUUUUGAUUGGGCUGCGAUAUGCCGAAAUUUAUUGGCCAAAAUAAAAUGGGCAAGACAUA .(((((((((..((((((....)).))))....))).))))))...(((((((.((((((.(((((((((.....))))..(((((......))))))))))))))))..))))).)).. ( -32.80) >DroSec_CAF1 9043 119 + 1 UUGCAUUGCUGGUGCGUUGGAAAAGUGCAAUAAUGCAAAUGCAUUUUGCUUGCAAUUUUGAUUUUGAUUGGGCUGUGAUAUGCCGAAAUUUAUUGGCCAAAAUAAAAUGGCCAACACAC- (((((..((.((((((((.......((((....))))))))))))..)).)))))............((.(((........))).)).....(((((((........))))))).....- ( -31.51) >DroSim_CAF1 11916 119 + 1 UUGCAUUGCUGGUGCGUUGGAAAAGUGCAAUAAUGCAAAUGCAUUUUGCUUGCAAUUUUGAUUUUGAUUGGGCUGUGAUAUGCCGAAAUUUAUUGGCCAAAAUAAAAUGGGCAACACAC- (((((..((.((((((((.......((((....))))))))))))..)).)))))............((.(((........))).)).....(((.(((........))).))).....- ( -26.91) >DroEre_CAF1 9619 95 + 1 -----UUGCAGGUGCGUUGGAAAAGUGCAAUAAUGCAAAUGCAUUUUGCUUGCAAUUUU------GAUUGAGUUGCGAUAUGCCGAAAUUUAUUGGCCAAACUAAA-------------- -----.((((..(((((((...........)))))))..)))).((((.(((((((((.------....)))))))))...(((((......))))))))).....-------------- ( -25.20) >DroYak_CAF1 10709 115 + 1 -----UUGCAGGUGCGUUGGAAAAGUGCAAUAAUGCAAAUGCAUUUUGCUUGCAAUUUUGAUUUUGAUUGCGUUGCGAUAUGCCAAAAUUUAUUGGCCAAAAUAAAAUGGCCAACAUAUG -----(((((..((((((....)).))))....)))))..((((.((((.(((((((........)))))))..)))).)))).........(((((((........)))))))...... ( -34.80) >consensus UUGCAUUGCAGGUGCGUUGGAAAAGUGCAAUAAUGCAAAUGCAUUUUGCUUGCAAUUUUGAUUUUGAUUGGGCUGCGAUAUGCCGAAAUUUAUUGGCCAAAAUAAAAUGGCCAACACAC_ ....(((((((...............((((.(((((....)))))))))...(((((........)))))..)))))))..(((((......)))))....................... (-19.36 = -19.52 + 0.16)


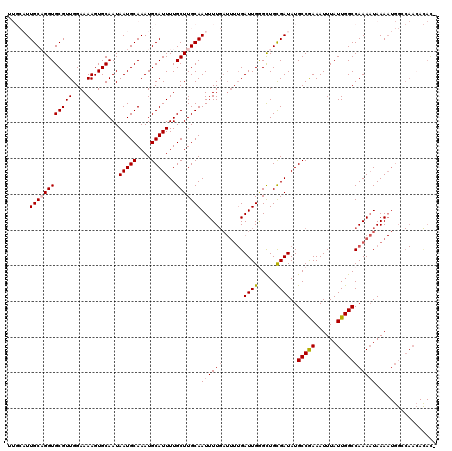
| Location | 8,548,104 – 8,548,224 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.58 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -14.74 |
| Energy contribution | -16.54 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8548104 120 - 22224390 UAUGUCUUGCCCAUUUUAUUUUGGCCAAUAAAUUUCGGCAUAUCGCAGCCCAAUCAAAAUCAAAAUUGCAAGCAAAAUGCAUUUGCAUUAUUGCACUUUUCCAACGCACCUGCAAUGCAA ..((.(((((..((((((((((((.........((.(((........))).))))))))).))))).)))))))...((((((((((....))))..........((....)))))))). ( -23.50) >DroSec_CAF1 9043 119 - 1 -GUGUGUUGGCCAUUUUAUUUUGGCCAAUAAAUUUCGGCAUAUCACAGCCCAAUCAAAAUCAAAAUUGCAAGCAAAAUGCAUUUGCAUUAUUGCACUUUUCCAACGCACCAGCAAUGCAA -...(((((((((........)))))))))......(((........)))...............(((((.(((((((((....))))).))))...........((....))..))))) ( -31.90) >DroSim_CAF1 11916 119 - 1 -GUGUGUUGCCCAUUUUAUUUUGGCCAAUAAAUUUCGGCAUAUCACAGCCCAAUCAAAAUCAAAAUUGCAAGCAAAAUGCAUUUGCAUUAUUGCACUUUUCCAACGCACCAGCAAUGCAA -.((((.((((...((((((......))))))....))))...))))..................(((((.(((((((((....))))).))))...........((....))..))))) ( -25.50) >DroEre_CAF1 9619 95 - 1 --------------UUUAGUUUGGCCAAUAAAUUUCGGCAUAUCGCAACUCAAUC------AAAAUUGCAAGCAAAAUGCAUUUGCAUUAUUGCACUUUUCCAACGCACCUGCAA----- --------------....(((.((((..........))).....((((.......------....))))..(((((((((....))))).))))......).)))((....))..----- ( -16.30) >DroYak_CAF1 10709 115 - 1 CAUAUGUUGGCCAUUUUAUUUUGGCCAAUAAAUUUUGGCAUAUCGCAACGCAAUCAAAAUCAAAAUUGCAAGCAAAAUGCAUUUGCAUUAUUGCACUUUUCCAACGCACCUGCAA----- ....(((((((((........))))))))).......(((....((...(((((..........)))))..(((((((((....))))).))))...........))...)))..----- ( -30.50) >consensus _AUGUGUUGCCCAUUUUAUUUUGGCCAAUAAAUUUCGGCAUAUCGCAGCCCAAUCAAAAUCAAAAUUGCAAGCAAAAUGCAUUUGCAUUAUUGCACUUUUCCAACGCACCUGCAAUGCAA ..(((((((..............(((..........)))......................((((.(((((....(((((....))))).))))).)))).)))))))............ (-14.74 = -16.54 + 1.80)



| Location | 8,548,144 – 8,548,259 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.24 |
| Mean single sequence MFE | -27.41 |
| Consensus MFE | -19.58 |
| Energy contribution | -19.27 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8548144 115 + 22224390 GCAUUUUGCUUGCAAUUUUGAUUUUGAUUGGGCUGCGAUAUGCCGAAAUUUAUUGGCCAAAAUAAAAUGGGCAAGACAUAU---GUAU--GUACGUGGCCUUGCUUCAAUUUAAUUAAUA (((.......)))....((((((((((..((((..((((((((.........(((.(((........))).))).......---))))--)).))..))))....))))...)))))).. ( -28.89) >DroSec_CAF1 9083 107 + 1 GCAUUUUGCUUGCAAUUUUGAUUUUGAUUGGGCUGUGAUAUGCCGAAAUUUAUUGGCCAAAAUAAAAUGGCCAACACAC-------------ACGUGGCCUUGCUUCAAUUUAAUUAAUA (((.......)))....((((((..((((((...(..(...((((.......(((((((........))))))).....-------------...)))).)..).)))))).)))))).. ( -24.06) >DroSim_CAF1 11956 107 + 1 GCAUUUUGCUUGCAAUUUUGAUUUUGAUUGGGCUGUGAUAUGCCGAAAUUUAUUGGCCAAAAUAAAAUGGGCAACACAC-------------ACGUGGCCUUGCUUCAAUUUAAUUAAUA (((.......)))....((((((((((..((((..((....(((((......)))))..........(((....).)).-------------.))..))))....))))...)))))).. ( -22.70) >DroYak_CAF1 10744 120 + 1 GCAUUUUGCUUGCAAUUUUGAUUUUGAUUGCGUUGCGAUAUGCCAAAAUUUAUUGGCCAAAAUAAAAUGGCCAACAUAUGUAACGUAUGUAUACUUGGCCUUGCUGCAAUUUAAUUAAUA (((.......)))....((((((..(((((((..((((...(((((.......((((((........))))))(((((((...)))))))....))))).))))))))))).)))))).. ( -34.00) >consensus GCAUUUUGCUUGCAAUUUUGAUUUUGAUUGGGCUGCGAUAUGCCGAAAUUUAUUGGCCAAAAUAAAAUGGCCAACACAC_____________ACGUGGCCUUGCUUCAAUUUAAUUAAUA (((.......)))....((((((..((((((...((((...((((.......(((((((........))))))).....................)))).)))).)))))).)))))).. (-19.58 = -19.27 + -0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:25 2006