
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,543,881 – 8,544,072 |
| Length | 191 |
| Max. P | 0.765739 |

| Location | 8,543,881 – 8,543,992 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.22 |
| Mean single sequence MFE | -26.52 |
| Consensus MFE | -24.71 |
| Energy contribution | -24.53 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8543881 111 - 22224390 UUUCCAGUUGCAGCGGCAUGUGGAAAAUUAGCGCUCAAUGUUAACAUUUGCCCACAUUGUUGCGUUGUGUUAAUGUAUUUUAAUUUUCCUCAGCCUCGAGUCCUCUGACAU--------- ....(((..((.(.(((.((.((((((((((......((((((((((.(((..........)))..))))))))))...)))))))))).))))).)..))...)))....--------- ( -26.90) >DroSec_CAF1 8074 111 - 1 UUUCCAGUUGCAGCGGCAUGUGGAAAAUUAGCGCUCAAUGUUAACAUUUGCCCACAUUGUUGCGUUGUGUUAAUGUAUUUUAAUUUUCCUCAGCCUCGAGUUCUCUGACAU--------- ....(((..((.(.(((.((.((((((((((......((((((((((.(((..........)))..))))))))))...)))))))))).))))).)..))...)))....--------- ( -27.60) >DroEre_CAF1 8100 111 - 1 UUUCCAGUUGCAGCGGCAUGUGGAAAAUUAGCGCUCAAUGUUAACAUUUGCCCACAUUGUUGCGUUGUGUUAAUGCAUUUUAAUUUUCCUCAGCCUCAAAUCCUCUGACAU--------- ....(((.....(.(((.((.(((((((((((((.((((((...(....)...))))))..)))).((((....))))..))))))))).))))).).......)))....--------- ( -27.50) >DroYak_CAF1 9856 120 - 1 UUUCCAGUUGCAGCGGCAUGUGGAAAAUUAGCGCUCAAUGUUAACAUUUGCCCACAUUGUUGCGUUGUGUUAAUGCAUUUUAAUUAUUCUCAGCCUCAAGUCCUCUGACAUUAUCUGAAA ....(((.(((((.(((.((.((......(((((.((((((...(....)...))))))..)))))((((....))))...............)).)).).)).))).))....)))... ( -24.10) >consensus UUUCCAGUUGCAGCGGCAUGUGGAAAAUUAGCGCUCAAUGUUAACAUUUGCCCACAUUGUUGCGUUGUGUUAAUGCAUUUUAAUUUUCCUCAGCCUCAAGUCCUCUGACAU_________ ....(((.....(.(((.((.(((((((((((((.((((((...(....)...))))))..)))).(((......)))..))))))))).))))).).......)))............. (-24.71 = -24.53 + -0.19)



| Location | 8,543,912 – 8,544,032 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.97 |
| Mean single sequence MFE | -35.05 |
| Consensus MFE | -31.64 |
| Energy contribution | -31.83 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8543912 120 + 22224390 AAAUACAUUAACACAACGCAACAAUGUGGGCAAAUGUUAACAUUGAGCGCUAAUUUUCCACAUGCCGCUGCAACUGGAAAAUGUGGAGGCUGCUCCGGUGCAGUGAAAACGGCUGGGGAA ...........(((..(((..((((((..((....))..)))))).)))...((((((((..(((....)))..))))))))))).......(((((((...((....)).))))))).. ( -34.90) >DroSec_CAF1 8105 120 + 1 AAAUACAUUAACACAACGCAACAAUGUGGGCAAAUGUUAACAUUGAGCGCUAAUUUUCCACAUGCCGCUGCAACUGGAAAAUGCGGAGGCUGUUCCGGUGCAGUGAAAACGGCUGGGGAA ................(((..((((((..((....))..)))))).))).....(((((.((.(((((((((.((((((...((....))..)))))))))))......))))))))))) ( -38.30) >DroEre_CAF1 8131 120 + 1 AAAUGCAUUAACACAACGCAACAAUGUGGGCAAAUGUUAACAUUGAGCGCUAAUUUUCCACAUGCCGCUGCAACUGGAAAAUGUGGAGGUUGUUUCGGAGCAGUGAAAACGGCUGGGGAA ............(((((((..((((((..((....))..)))))).)).......((((((((.(((.......)))...)))))))))))))(((..(((.((....)).)))..))). ( -32.60) >DroYak_CAF1 9896 120 + 1 AAAUGCAUUAACACAACGCAACAAUGUGGGCAAAUGUUAACAUUGAGCGCUAAUUUUCCACAUGCCGCUGCAACUGGAAAAUGUGCAGGCUUUUCCGGAGCAGGGAAAACGGCUGGGGAA ................(((..((((((..((....))..)))))).))).....(((((.((.((((((((..((((((((.((....)))))))))).))))......))))))))))) ( -34.40) >consensus AAAUACAUUAACACAACGCAACAAUGUGGGCAAAUGUUAACAUUGAGCGCUAAUUUUCCACAUGCCGCUGCAACUGGAAAAUGUGGAGGCUGUUCCGGAGCAGUGAAAACGGCUGGGGAA ................(((..((((((..((....))..)))))).))).....(((((.((.(((((((((.((((((...((....))..)))))))))))......))))))))))) (-31.64 = -31.83 + 0.19)

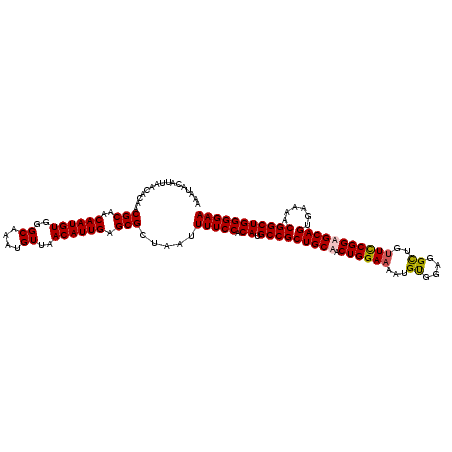
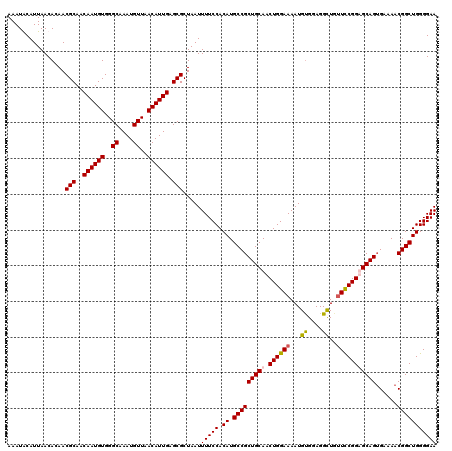
| Location | 8,543,952 – 8,544,072 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.03 |
| Mean single sequence MFE | -34.55 |
| Consensus MFE | -29.71 |
| Energy contribution | -30.15 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8543952 120 + 22224390 CAUUGAGCGCUAAUUUUCCACAUGCCGCUGCAACUGGAAAAUGUGGAGGCUGCUCCGGUGCAGUGAAAACGGCUGGGGAAAAUACGCGUGGCAAGUGAGAGACAGAUGACAAAGGAAAUU ((((..((((..(((((((.((.(((((((((.(((((....(..(...)..)))))))))))......)))))).)))))))..))))....))))....................... ( -34.70) >DroSec_CAF1 8145 120 + 1 CAUUGAGCGCUAAUUUUCCACAUGCCGCUGCAACUGGAAAAUGCGGAGGCUGUUCCGGUGCAGUGAAAACGGCUGGGGAAAAUAAGAGUGGCAAGUGAGAGGCAGAUGACAAAGGAAAUU ((((...((((.(((((((.((.(((((((((.((((((...((....))..)))))))))))......)))))).)))))))...))))...))))....................... ( -37.20) >DroEre_CAF1 8171 120 + 1 CAUUGAGCGCUAAUUUUCCACAUGCCGCUGCAACUGGAAAAUGUGGAGGUUGUUUCGGAGCAGUGAAAACGGCUGGGGAAAAUACGAGUGGCAAGUGAGAGACAGAUGACAAAAGAAAUU ..(((....((...(((((((.(((((((((((((............))))))(((..(((.((....)).)))..))).......))))))).))).)))).))....)))........ ( -28.70) >DroYak_CAF1 9936 120 + 1 CAUUGAGCGCUAAUUUUCCACAUGCCGCUGCAACUGGAAAAUGUGCAGGCUUUUCCGGAGCAGGGAAAACGGCUGGGGAAAAUGCGAGUGGCAAGUGAGAGACAGAUGACAAUGGAAAUU (((((....((...(((((((.((((((((((.((((((((.((....))))))))))(((.(......).)))........))).))))))).))).)))).))....)))))...... ( -37.60) >consensus CAUUGAGCGCUAAUUUUCCACAUGCCGCUGCAACUGGAAAAUGUGGAGGCUGUUCCGGAGCAGUGAAAACGGCUGGGGAAAAUACGAGUGGCAAGUGAGAGACAGAUGACAAAGGAAAUU ((((...((((.(((((((.((.(((((((((.((((((...((....))..)))))))))))......)))))).)))))))...))))...))))....................... (-29.71 = -30.15 + 0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:19 2006