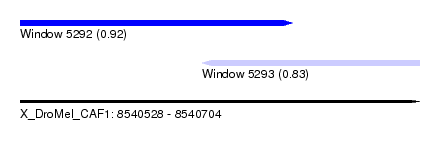
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,540,528 – 8,540,704 |
| Length | 176 |
| Max. P | 0.919985 |

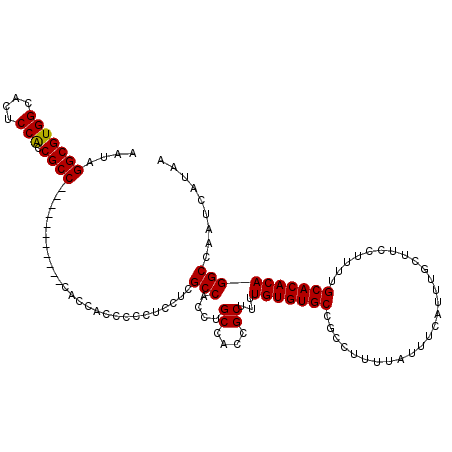
| Location | 8,540,528 – 8,540,648 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.76 |
| Mean single sequence MFE | -22.46 |
| Consensus MFE | -21.33 |
| Energy contribution | -21.17 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8540528 120 + 22224390 AAUAGGCGUGGCACUCCACCGCCCAGCAGCCACCCACCACCCCCUCCUCGCCACCUGCCAACGCUUUUGUGUGCCGCCUUUUAUUUCAUUUGCUUCCUUUUGCACACAGGCCAAUCAUAA ....((((((((((.(....((...((((.........................))))....))....).))))))).............(((........))).....)))........ ( -23.31) >DroSec_CAF1 4769 109 + 1 AAUAGGCGUGGCACUCCACCGCC-----------CACCACCCUCUCAUCGCCACCUGCUACCGCUUUUGUGUGCCGCCUUUUAUUUCAUUUGCUUCCUUUUGCACACAGGCCAAUCAUAA ....(((((((....))).))))-----------...............(((....((....))...(((((((...........................))))))))))......... ( -22.13) >DroSim_CAF1 7308 109 + 1 AAUAGGCGUGGCACUCCACCGCC-----------CACCACCCUCUCCUCGCCACCUGCCACCGCUUUUGUGUGCCGCCUUUUAUUUCAUUUGCUUCCUUUUGCACACAGGCCAAUCAUAA ....(((((((....))).))))-----------...............(((....((....))...(((((((...........................))))))))))......... ( -21.63) >DroEre_CAF1 4740 112 + 1 AAUAGGCGUGGCACUCCACCGCCC--------CCGACCACCCCCUCCACGCCACCUGCCGUCGCUUUUGUGUGCCGCCUUUUAUUUCAUUUGCUUCCUUUUGCACACAGGCCAAUCAUAA ....(((((((........((...--------.))..........)))))))..........(((..(((((((...........................))))))))))......... ( -22.10) >DroYak_CAF1 5712 102 + 1 AAUAGGCGUGGCACUCCGCCGCC------------------CCCUCCUCGCCACCUGCCGUCGCUUUUGUGUGCCGCCUUUUAUUUCAUUUGCUUCCUUUUGCACACAGGCCAAUCAUAA ....((.(((((.....))))))------------------).......(((....((....))...(((((((...........................))))))))))......... ( -23.13) >consensus AAUAGGCGUGGCACUCCACCGCC___________CACCACCCCCUCCUCGCCACCUGCCACCGCUUUUGUGUGCCGCCUUUUAUUUCAUUUGCUUCCUUUUGCACACAGGCCAAUCAUAA ....(((((((....))).))))..........................(((....((....))...(((((((...........................))))))))))......... (-21.33 = -21.17 + -0.16)


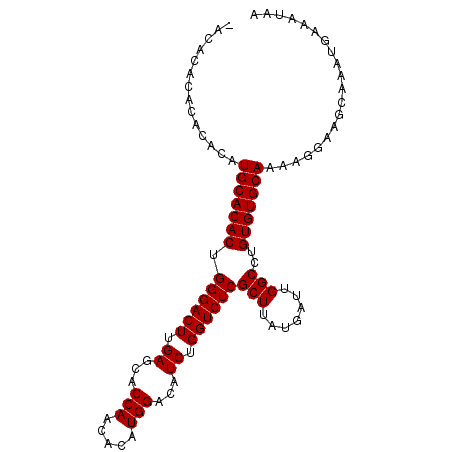
| Location | 8,540,608 – 8,540,704 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 97.30 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -24.60 |
| Energy contribution | -24.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8540608 96 - 22224390 ---ACAAACACACAUGCACACUGGGACUUGAGCACCAACACAUGGACAUCUGGUCUCGCUUAUGAUUGGCCUGUGUGCAAAAGGAAGCAAAUGAAAUAA ---...........(((((((.((((((.((...(((.....)))...)).))))))(((.......)))..))))))).................... ( -24.60) >DroSec_CAF1 4838 98 - 1 -ACACACACACACAUGCACACUGGGACUUGAGCACCAACACAUGGACAUCUGGUCUCGCUUAUGAUUGGCCUGUGUGCAAAAGGAAGCAAAUGAAAUAA -.............(((((((.((((((.((...(((.....)))...)).))))))(((.......)))..))))))).................... ( -24.60) >DroSim_CAF1 7377 99 - 1 CACACACACACACAUGCACACUGGGACUUGAGCACCAACACAUGGACAUCUGGUCUCGCUUAUGAUUGGCCUGUGUGCAAAAGGAAGCAAAUGAAAUAA ..............(((((((.((((((.((...(((.....)))...)).))))))(((.......)))..))))))).................... ( -24.60) >consensus _ACACACACACACAUGCACACUGGGACUUGAGCACCAACACAUGGACAUCUGGUCUCGCUUAUGAUUGGCCUGUGUGCAAAAGGAAGCAAAUGAAAUAA ..............(((((((.((((((.((...(((.....)))...)).))))))(((.......)))..))))))).................... (-24.60 = -24.60 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:15 2006