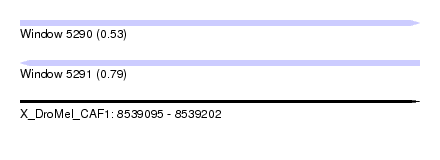
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,539,095 – 8,539,202 |
| Length | 107 |
| Max. P | 0.785647 |

| Location | 8,539,095 – 8,539,202 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.36 |
| Mean single sequence MFE | -37.02 |
| Consensus MFE | -28.24 |
| Energy contribution | -28.44 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8539095 107 + 22224390 AGGUUGAUUUCGU-----UGCUUUUCGCUGAGCGUUUGGCCAGCGACUGUGGAAAUGCAGGCAGCAUCCUGCGGCUUUUAUGCAGUUGCCAAAGGACUCGGGCUAAUAUGU--------A ((.((((....((-----(((...((((((.((.....))))))))((((......))))))))).((((..(((............)))..)))).)))).)).......--------. ( -34.50) >DroSec_CAF1 3327 107 + 1 AGCUUGAUUUCGU-----UGCUUUUCGCUGAGCGUUUGGCCAGCGACUGUGGAAAUGCAGGCAGCAUCCUGCGGCUUUUAUGCAGUUGCCAAAGGACUCGGGCUAAUAUGU--------A (((((((....((-----(((...((((((.((.....))))))))((((......))))))))).((((..(((............)))..)))).))))))).......--------. ( -39.00) >DroSim_CAF1 5848 107 + 1 AGCUUGAUUUCCU-----UGCUUUUCGCUGAGCGUUUGGCCCGCGACUGUGGAAAUGCAGGCAGCAUCCUGCGGCUUUUAUGCAAUUGCCAAAGGACUCGAGCUAAUAUGU--------A (((((((..((((-----(((((......))))...((((((((....))))...((((((......))))))((......))....))))))))).))))))).......--------. ( -36.60) >DroEre_CAF1 3295 120 + 1 AAGUUUAUUUCGUAGAGUUGCUUUUCGCUGAGCGUUUGGCCAGCGACUGUGGAAAUGCAGGCAGCAUCCUGCGGCUUUUAUGCAGCUGGCAAAGGACUCGGGCUAAUAUAUACGUAUGUA .....(((..(((((((......))).((((((.((((.(((((..(.((((((.((((((......))))))..)))))))..))))))))).).))))).........))))...))) ( -36.20) >DroYak_CAF1 3453 112 + 1 AAGUUUAUUGCGUAGAGCUGCUUUUCGCUGAGCGUUUGGCCAGCGACUGUGGAAAUGCAGGCAGCAUCCUGCGGCUUUUAUGCAGCUGCCAAAGGACUCGGGCUAAUAUAU--------G .((((..(((.((((.(((((...((((((.((.....))))))))((((......)))))))))...(((((.......))))))))))))..)))).............--------. ( -38.80) >consensus AGGUUGAUUUCGU_____UGCUUUUCGCUGAGCGUUUGGCCAGCGACUGUGGAAAUGCAGGCAGCAUCCUGCGGCUUUUAUGCAGUUGCCAAAGGACUCGGGCUAAUAUGU________A ..........................(((((((.((((((.(((...(((((((.((((((......))))))..)))))))..))))))))).).))).)))................. (-28.24 = -28.44 + 0.20)

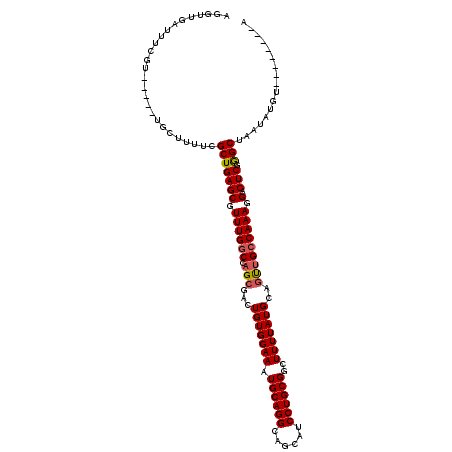
| Location | 8,539,095 – 8,539,202 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.36 |
| Mean single sequence MFE | -30.56 |
| Consensus MFE | -24.26 |
| Energy contribution | -24.50 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8539095 107 - 22224390 U--------ACAUAUUAGCCCGAGUCCUUUGGCAACUGCAUAAAAGCCGCAGGAUGCUGCCUGCAUUUCCACAGUCGCUGGCCAAACGCUCAGCGAAAAGCA-----ACGAAAUCAACCU .--------........(((.((....)).)))...(((.........(((((......)))))..........((((((((.....)).))))))...)))-----............. ( -27.50) >DroSec_CAF1 3327 107 - 1 U--------ACAUAUUAGCCCGAGUCCUUUGGCAACUGCAUAAAAGCCGCAGGAUGCUGCCUGCAUUUCCACAGUCGCUGGCCAAACGCUCAGCGAAAAGCA-----ACGAAAUCAAGCU .--------.......(((....(((((.((((............)))).))))))))((.((.(((((.....((((((((.....)).))))))......-----..))))))).)). ( -28.42) >DroSim_CAF1 5848 107 - 1 U--------ACAUAUUAGCUCGAGUCCUUUGGCAAUUGCAUAAAAGCCGCAGGAUGCUGCCUGCAUUUCCACAGUCGCGGGCCAAACGCUCAGCGAAAAGCA-----AGGAAAUCAAGCU .--------.......((((.((.(((((..((....))......((.(((((......)))))..........((((((((.....)))).))))...)))-----))))..)).)))) ( -30.80) >DroEre_CAF1 3295 120 - 1 UACAUACGUAUAUAUUAGCCCGAGUCCUUUGCCAGCUGCAUAAAAGCCGCAGGAUGCUGCCUGCAUUUCCACAGUCGCUGGCCAAACGCUCAGCGAAAAGCAACUCUACGAAAUAAACUU ......((((...........((((..((.((((((.((......)).(((((......)))))............)))))).))..)))).((.....)).....)))).......... ( -31.50) >DroYak_CAF1 3453 112 - 1 C--------AUAUAUUAGCCCGAGUCCUUUGGCAGCUGCAUAAAAGCCGCAGGAUGCUGCCUGCAUUUCCACAGUCGCUGGCCAAACGCUCAGCGAAAAGCAGCUCUACGCAAUAAACUU .--------........(((.((....)).)))((((((.........(((((......)))))..........((((((((.....)).))))))...))))))............... ( -34.60) >consensus U________ACAUAUUAGCCCGAGUCCUUUGGCAACUGCAUAAAAGCCGCAGGAUGCUGCCUGCAUUUCCACAGUCGCUGGCCAAACGCUCAGCGAAAAGCA_____ACGAAAUCAACCU .................(((.((....)).)))...(((.........(((((......)))))..........((((((((.....)).))))))...))).................. (-24.26 = -24.50 + 0.24)

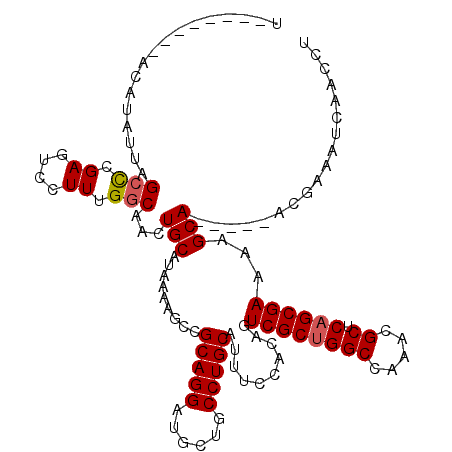
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:13 2006