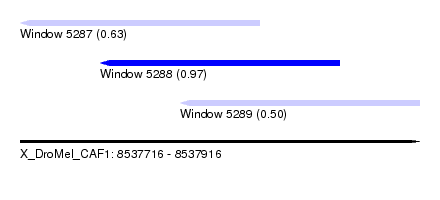
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,537,716 – 8,537,916 |
| Length | 200 |
| Max. P | 0.966514 |

| Location | 8,537,716 – 8,537,836 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.58 |
| Mean single sequence MFE | -48.00 |
| Consensus MFE | -40.12 |
| Energy contribution | -40.84 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8537716 120 - 22224390 UACGGCUCGACCGCCCAGCCAGACGCCGGAGAACCUCUCUUCCGCGAGCAAGGAUUCGGAACUGGCGGAGGUGGGCAAUGGACCCAAUUCGGGGAGCUUCACCAUGAUGCACCCGGCAUU ...(((((..((((((.(((((...(((((...((((((......)))..))).)))))..))))).).)))))........(((.....)))))))).......(((((.....))))) ( -44.60) >DroSec_CAF1 1944 120 - 1 UACGGCUCGACCGCCCAGCCAGACGCCGGAGAACCUCUCUUCCGCCAGCAAGGAUUCGGAGCUGGCGUAGGUGAGCAAUGGACCCAGUUCGGGGAACUUCACCAUGAUGCACCCGGUAUU ...((((.(.....).))))....(((((......((.(((.(((((((...........))))))).))).))(((((((.(((.....)))(.....).))))..)))..)))))... ( -40.30) >DroSim_CAF1 1962 120 - 1 UACGGCUCGACCGCCCAGCCAGACGCCGGAGAACCUCUCUUCCGCCAGCAAGGAUUCGGAGCUGGCGGAGGUGAGCAAUGGACCUAGUUCGGGGAACUUCACAAUGAUGCACCCGGCAUU ...((((.(.....).))))....(((((.....(((.(((((((((((...........))))))))))).)))((.((.....(((((...)))))...)).))......)))))... ( -45.80) >DroEre_CAF1 2034 120 - 1 UCCGGGUCGACCGCCCAGCCAGACGCCGGAGAACCUCUCCUCCGCCAGCAAGGAUUCCGAUCUGGCGGAGGUAAGCAAUGGACCCAGUGCGGGCAGCUUCACCAUGAUGCACCCGGCAUU .((((((.....((((........(((((((.....))))((((((((...((...))...)))))))))))..(((.((....)).))))))).((.((.....)).)))))))).... ( -53.10) >DroYak_CAF1 2022 120 - 1 UCCGGCUCGACCGCCCAGCCAAACGCCGGAGAAUCUCUCCUCCGCCAGCAAGGAUUCCGAGCUGGCGGAGGUGAGUAAUGGACCCAGUGCGGGGAGCUUCACCAUGAUGCACCCGGCAUU (((((((.(.....).))))...((((((((.....))))(((((((((..((...))..)))))))))))))......)))...(((((.(((.((.((.....)).)).))).))))) ( -56.20) >consensus UACGGCUCGACCGCCCAGCCAGACGCCGGAGAACCUCUCUUCCGCCAGCAAGGAUUCGGAGCUGGCGGAGGUGAGCAAUGGACCCAGUUCGGGGAGCUUCACCAUGAUGCACCCGGCAUU ...((((.(.....).))))....(((((......((.(((((((((((...........))))))))))).))(((((((.(((.....)))(.....).))))..)))..)))))... (-40.12 = -40.84 + 0.72)


| Location | 8,537,756 – 8,537,876 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.42 |
| Mean single sequence MFE | -59.90 |
| Consensus MFE | -49.38 |
| Energy contribution | -50.82 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8537756 120 - 22224390 UGGGCGGUGGAGCGGUGCAGGGCGGAUCCGGCAACGGAGCUACGGCUCGACCGCCCAGCCAGACGCCGGAGAACCUCUCUUCCGCGAGCAAGGAUUCGGAACUGGCGGAGGUGGGCAAUG ((((((((.((((.((....(((...((((....))))))))).)))).))))))))(((...((((((((.....))))(((((.((.............)).)))))))))))).... ( -59.72) >DroSec_CAF1 1984 120 - 1 UGGGCGGCGGAGCAGUGCAGGGGGGAUCCGGCAACGGGGCUACGGCUCGACCGCCCAGCCAGACGCCGGAGAACCUCUCUUCCGCCAGCAAGGAUUCGGAGCUGGCGUAGGUGAGCAAUG ...((((((((((...))(((((((.((((((..(..((((..(((......))).)))).)..))))))...))))))))))))).))...........(((.((....)).))).... ( -52.80) >DroSim_CAF1 2002 120 - 1 UGGGCGGCGGAGCAGUGCAGGGGGGAUCCGGCAACGGGGCUACGGCUCGACCGCCCAGCCAGACGCCGGAGAACCUCUCUUCCGCCAGCAAGGAUUCGGAGCUGGCGGAGGUGAGCAAUG ....(((((...(.(.((.(((.((.((((....))((((....)))))))).))).))).).)))))......(((.(((((((((((...........))))))))))).)))..... ( -58.30) >DroEre_CAF1 2074 120 - 1 UGGGCGGCGGAGCGGUACAGGGGGGAUCCGGCAACGGUGCUCCGGGUCGACCGCCCAGCCAGACGCCGGAGAACCUCUCCUCCGCCAGCAAGGAUUCCGAUCUGGCGGAGGUAAGCAAUG ...(((((((.(((((((..((((.(.(((....)))).))))..))..))))))).)))....(((((((.....))))((((((((...((...))...)))))))))))..)).... ( -59.00) >DroYak_CAF1 2062 120 - 1 UGGGCGGCGGAGCGGUGCAGGGGGCGUCCGGCAACGGAGCUCCGGCUCGACCGCCCAGCCAAACGCCGGAGAAUCUCUCCUCCGCCAGCAAGGAUUCCGAGCUGGCGGAGGUGAGUAAUG ..((((((((.(((((((..(((((.((((....))))))))).))...))))))).)))....))).......(((.(((((((((((..((...))..))))))))))).)))..... ( -69.70) >consensus UGGGCGGCGGAGCGGUGCAGGGGGGAUCCGGCAACGGAGCUACGGCUCGACCGCCCAGCCAGACGCCGGAGAACCUCUCUUCCGCCAGCAAGGAUUCGGAGCUGGCGGAGGUGAGCAAUG (((((((..((((.((((........((((....)))))).)).))))..)))))))((....((((((((.....))))(((((((((...........))))))))))))).)).... (-49.38 = -50.82 + 1.44)



| Location | 8,537,796 – 8,537,916 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -63.14 |
| Consensus MFE | -55.98 |
| Energy contribution | -56.38 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8537796 120 - 22224390 UGGCGGAUGCGCUGCCUCAGCCUGGGCACGCCCCCCGUCAUGGGCGGUGGAGCGGUGCAGGGCGGAUCCGGCAACGGAGCUACGGCUCGACCGCCCAGCCAGACGCCGGAGAACCUCUCU .((((..((((((((..((((....)).(((((........))))).))..))))))))((((((.((((....))((((....)))))))))))).......))))((((.....)))) ( -65.80) >DroSec_CAF1 2024 120 - 1 UGGCGGAUGCGCUGCCUCAGCCUGGGCACGCCUCCCGUCAUGGGCGGCGGAGCAGUGCAGGGGGGAUCCGGCAACGGGGCUACGGCUCGACCGCCCAGCCAGACGCCGGAGAACCUCUCU .((((..((((((((.((.((....)).((((.(((.....))).))))))))))))))(((.((.((((....))((((....)))))))).))).......))))((((.....)))) ( -63.40) >DroSim_CAF1 2042 120 - 1 UGGCGGAUGCGCUGCCUCAGCCUGGGAACGCCUCCCGUCAUGGGCGGCGGAGCAGUGCAGGGGGGAUCCGGCAACGGGGCUACGGCUCGACCGCCCAGCCAGACGCCGGAGAACCUCUCU .((((..(((((((((((.....)))..((((.(((.....))).))))..))))))))(((.((.((((....))((((....)))))))).))).......))))((((.....)))) ( -60.60) >DroEre_CAF1 2114 120 - 1 UGGCGGAUGCGGUGCCUCAGCCUGGGCACGCCCCCCGUCAUGGGCGGCGGAGCGGUACAGGGGGGAUCCGGCAACGGUGCUCCGGGUCGACCGCCCAGCCAGACGCCGGAGAACCUCUCC .((((..(((.((((((......))))))(((.(((.....))).)))((.(((((((..((((.(.(((....)))).))))..))..))))))).).))..))))((((.....)))) ( -60.60) >DroYak_CAF1 2102 120 - 1 UGGCGGAUGCGCUGCCUCAGCCUGGGCACGCCCCCCGUCAUGGGCGGCGGAGCGGUGCAGGGGGCGUCCGGCAACGGAGCUCCGGCUCGACCGCCCAGCCAAACGCCGGAGAAUCUCUCC .((((..((((((((.((.((....)).((((.(((.....))).)))))))))))))).(((((.((((....)))))))))((((.(.....).))))...))))((((.....)))) ( -65.30) >consensus UGGCGGAUGCGCUGCCUCAGCCUGGGCACGCCCCCCGUCAUGGGCGGCGGAGCGGUGCAGGGGGGAUCCGGCAACGGAGCUACGGCUCGACCGCCCAGCCAGACGCCGGAGAACCUCUCU .((((..((((((((.((.((....)).((((.(((.....))).))))))))))))))(((.((.((((....))((((....)))))))).))).......))))((((.....)))) (-55.98 = -56.38 + 0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:11 2006