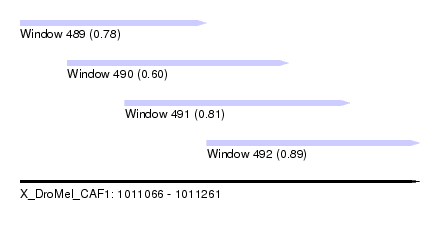
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,011,066 – 1,011,261 |
| Length | 195 |
| Max. P | 0.892975 |

| Location | 1,011,066 – 1,011,157 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -26.12 |
| Energy contribution | -25.90 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1011066 91 + 22224390 AGGCGCCAGGUGCCAGGUGCCAGGGGCCCGGGUCAAGGGUCAAGAAGGCUUAGUACAGCGUGAGUAGGCUUAAAAUGUAUCUGCACAAAUU .((((((.(....).))))))...(((((.......)))))......((...(((((...((((....))))...)))))..))....... ( -30.20) >DroSec_CAF1 96542 82 + 1 AG-------GUGCCAGGUGCCAGGGACCCGGGUCAAGGGUCAAGAAGGCUUAG--CAGCGUGAGUAGGCUUAAAAUGUAUCUGCACAAACU ..-------((((.((((((....(((((.......)))))....((((((..--..((....)))))))).....))))))))))..... ( -28.40) >DroSim_CAF1 73740 82 + 1 AG-------GUGCCAGGUGCCAGGGACCCGGGUCAAGGGUCAAGAAGGCUUAG--CAGCGUGAGUAGGCUUAAAAUGUAUCUGCACAAACU ..-------((((.((((((....(((((.......)))))....((((((..--..((....)))))))).....))))))))))..... ( -28.40) >consensus AG_______GUGCCAGGUGCCAGGGACCCGGGUCAAGGGUCAAGAAGGCUUAG__CAGCGUGAGUAGGCUUAAAAUGUAUCUGCACAAACU .........((((.((((((....(((((.......)))))(((...(((((........)))))...))).....))))))))))..... (-26.12 = -25.90 + -0.22)

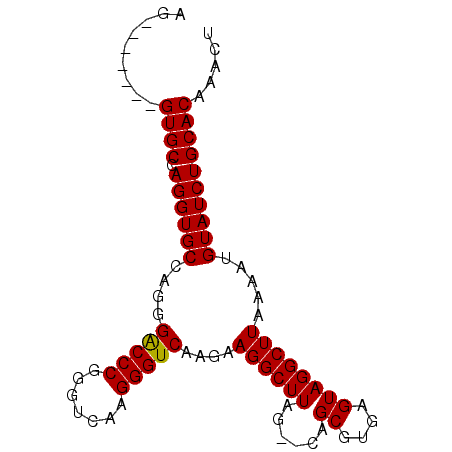
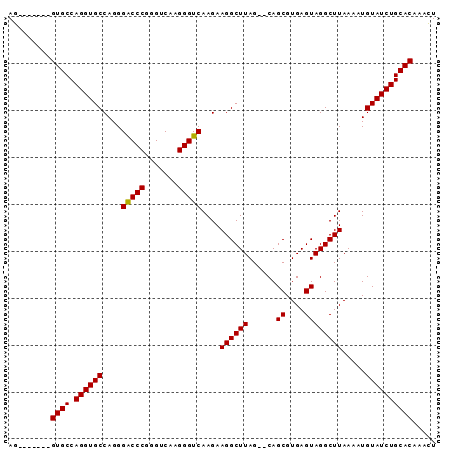
| Location | 1,011,089 – 1,011,197 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 96.27 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -30.59 |
| Energy contribution | -29.93 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.37 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1011089 108 + 22224390 GGGCCCGGGUCAAGGGUCAAGAAGGCUUAGUACAGCGUGAGUAGGCUUAAAAUGUAUCUGCACAAAUUUCAAAUACGAACACAAGAGCACAGACACGCAGUGUUUGUG .(((((.......)))))......((...(((((...((((....))))...)))))..))..........................(((((((((...))))))))) ( -27.50) >DroSec_CAF1 96558 106 + 1 GGACCCGGGUCAAGGGUCAAGAAGGCUUAG--CAGCGUGAGUAGGCUUAAAAUGUAUCUGCACAAACUUCAAAUACGAACACAAGAGCACAGACACGCAGUGUCUGUG .(((((.......)))))..(((((((...--.)))(((((...((.......))..)).)))...)))).................(((((((((...))))))))) ( -31.70) >DroSim_CAF1 73756 106 + 1 GGACCCGGGUCAAGGGUCAAGAAGGCUUAG--CAGCGUGAGUAGGCUUAAAAUGUAUCUGCACAAACUUCAAAUACGAACACGAGAGCACAGACACGCAGUGUCUGUG .(((((.......)))))..(((((((...--.)))(((((...((.......))..)).)))...)))).................(((((((((...))))))))) ( -31.70) >consensus GGACCCGGGUCAAGGGUCAAGAAGGCUUAG__CAGCGUGAGUAGGCUUAAAAUGUAUCUGCACAAACUUCAAAUACGAACACAAGAGCACAGACACGCAGUGUCUGUG .(((((.......)))))..(((((((......)))(((((...((.......))..)).)))...)))).................(((((((((...))))))))) (-30.59 = -29.93 + -0.66)

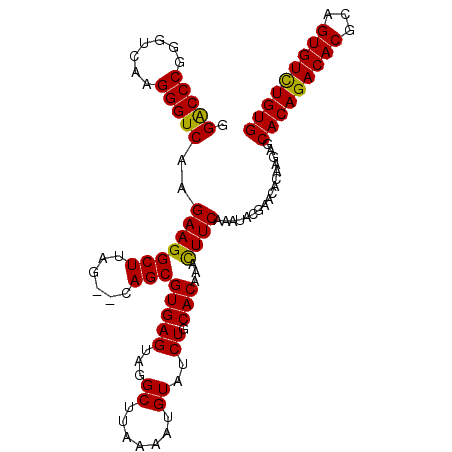
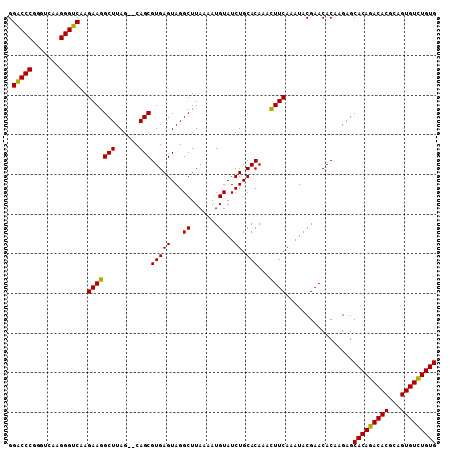
| Location | 1,011,117 – 1,011,227 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -31.73 |
| Consensus MFE | -25.94 |
| Energy contribution | -25.50 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1011117 110 + 22224390 AGUACAGCGUGAGUAGGCUUAAAAUGUAUCUGCACAAAUUUCAAAUACGAACACAAGAGCACAGACACGCAGUGUUUGUGUGGCCAAAAGGGCCCAAAGUCGAAGAUAUA (((...((....))..))).....(((((((..((.......................((((((((((...))))))))))((((.....))))....))...))))))) ( -28.40) >DroSec_CAF1 96586 107 + 1 AG--CAGCGUGAGUAGGCUUAAAAUGUAUCUGCACAAACUUCAAAUACGAACACAAGAGCACAGACACGCAGUGUCUGUGUGGCCAAAAGGGC-CAAAGUCGAAGAUAUA ((--(.((....))..))).....(((((((..((.......................((((((((((...))))))))))((((.....)))-)...))...))))))) ( -33.00) >DroSim_CAF1 73784 107 + 1 AG--CAGCGUGAGUAGGCUUAAAAUGUAUCUGCACAAACUUCAAAUACGAACACGAGAGCACAGACACGCAGUGUCUGUGUGGCCAAAAGGGC-CAAAGUCGAAGAUAUA ((--(.((....))..))).....(((((((........(((......)))..(((..((((((((((...))))))))))((((.....)))-)....))).))))))) ( -33.80) >consensus AG__CAGCGUGAGUAGGCUUAAAAUGUAUCUGCACAAACUUCAAAUACGAACACAAGAGCACAGACACGCAGUGUCUGUGUGGCCAAAAGGGC_CAAAGUCGAAGAUAUA ........(((((...((.......))..)).)))...((((................((((((((((...))))))))))((((.....))).)......))))..... (-25.94 = -25.50 + -0.44)

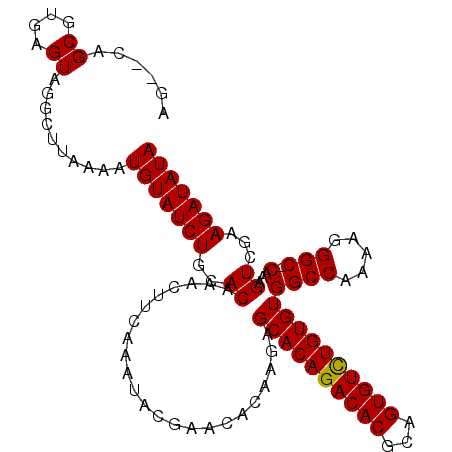
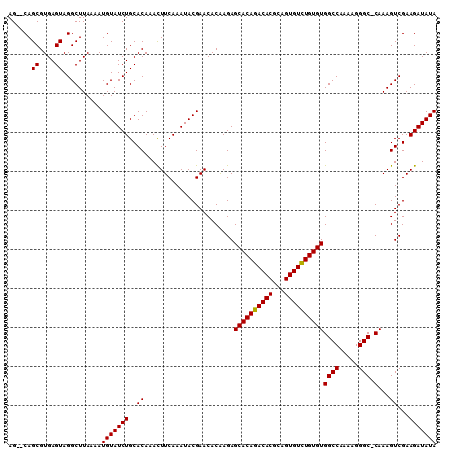
| Location | 1,011,157 – 1,011,261 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 98.07 |
| Mean single sequence MFE | -32.25 |
| Consensus MFE | -28.97 |
| Energy contribution | -28.75 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1011157 104 + 22224390 UCAAAUACGAACACAAGAGCACAGACACGCAGUGUUUGUGUGGCCAAAAGGGCCCAAAGUCGAAGAUAUAGCCGCACUUUGCAAAUCGUGACUGGGCAAAGGCA .......(((........((((((((((...))))))))))((((.....)))).....)))...........((.((((((.............)))))))). ( -29.92) >DroSec_CAF1 96624 103 + 1 UCAAAUACGAACACAAGAGCACAGACACGCAGUGUCUGUGUGGCCAAAAGGGC-CAAAGUCGAAGAUAUAGCCGCACUUUGCAAAUCGUGACUGGGCAAAGGCA .......(((........((((((((((...))))))))))((((.....)))-)....)))...........((.((((((.............)))))))). ( -33.42) >DroSim_CAF1 73822 103 + 1 UCAAAUACGAACACGAGAGCACAGACACGCAGUGUCUGUGUGGCCAAAAGGGC-CAAAGUCGAAGAUAUAGCCGCACUUUGCAAAUCGUGACUGGGCAAAGGCA .............(((..((((((((((...))))))))))((((.....)))-)....)))...........((.((((((.............)))))))). ( -33.42) >consensus UCAAAUACGAACACAAGAGCACAGACACGCAGUGUCUGUGUGGCCAAAAGGGC_CAAAGUCGAAGAUAUAGCCGCACUUUGCAAAUCGUGACUGGGCAAAGGCA .......(((........((((((((((...))))))))))((((.....))).)....)))...........((.((((((.............)))))))). (-28.97 = -28.75 + -0.22)

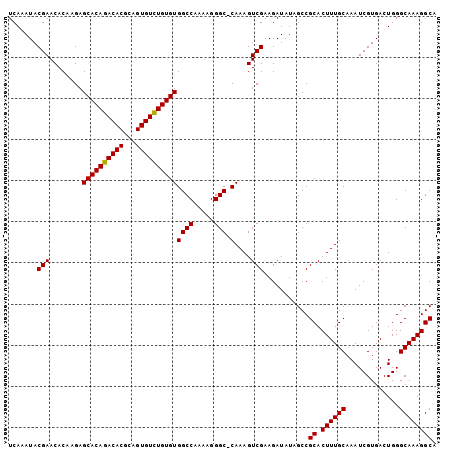
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:44 2006