| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,527,072 – 8,527,172 |
| Length | 100 |
| Max. P | 0.922800 |

| Location | 8,527,072 – 8,527,172 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.07 |
| Mean single sequence MFE | -27.83 |
| Consensus MFE | -16.09 |
| Energy contribution | -17.63 |
| Covariance contribution | 1.54 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8527072 100 + 22224390 -----------UGCCAGCGAAAUA---GGAAGAUGGCACAAAAACUUCCGAAACUUCGCUCUUCCUCUUCACG-----CCACACACACACAUGCGC-AAAUUAGGAGUGCUCGUGUGUGU -----------....((((((...---(((((.(........).))))).....)))))).............-----....((((((((..((((-.........))))..)))))))) ( -28.10) >DroSec_CAF1 1971 100 + 1 -----------UGCCAGCGAAAUA---GGAAGAUGACAUAAAAACUUCCGAAACUUCGCUCUUCCUCUUCACG-----CCACACACACACAUGCGC-AAAUUAGGAGUGCUCGUGAGUGU -----------....((((((...---(((((.(........).))))).....)))))).............-----....((((.(((..((((-.........))))..))).)))) ( -23.40) >DroSim_CAF1 1974 100 + 1 -----------UGCCAGCGAAAUA---GGAAGAUGACACAAAAACUUCCGAAACUUCGCUCUUCCUCUUCACG-----CCACACACACACAUGCGC-AAAUUAGGAGUGCUCGUGUGUGU -----------....((((((...---(((((.(........).))))).....)))))).............-----....((((((((..((((-.........))))..)))))))) ( -28.10) >DroEre_CAF1 1913 95 + 1 -----------UGCCAGCGAAAUA---GGAAGAUGACACAAAAACUUCCGAAACUUCGCUCUUCCUCUUCGCU-----CCACA--CACACAUGCGC-AAAUUAGGAGUGCUCGUGUG--- -----------....((((((..(---(((((((((...................))).))))))).))))))-----.....--(((((..((((-.........))))..)))))--- ( -25.71) >DroYak_CAF1 1911 96 + 1 -----------UUCCAGCGAAAUA---GGAAGAUGACACAAAAACUUCCGAAACUUCGCUCUUCUUCUACACU-----CCACA--CACACAUGCGC-AAAUUAGGAGUGUUCGUGUGU-- -----------....((((((...---(((((.(........).))))).....)))))).............-----....(--(((((..((((-.........))))..))))))-- ( -22.20) >DroPer_CAF1 38567 118 + 1 CUUAUACAUUAUACCAACAAAAAAAAUGGGAGAUGAGACUGCUGCUUCCGUCUCGUCU--CCCUCUCUUCGCGCUUAUCCAUACACAAACAUGCGCGAAAUUAACUGUAGUUGUGUGUGC .........(((((((((.........((((((((((((..........)))))))))--)))....(((((((..................)))))))..........)))).))))). ( -39.47) >consensus ___________UGCCAGCGAAAUA___GGAAGAUGACACAAAAACUUCCGAAACUUCGCUCUUCCUCUUCACG_____CCACACACACACAUGCGC_AAAUUAGGAGUGCUCGUGUGUGU ...............((((((......(((((............))))).....)))))).......................(((((((..((((..........))))..))))))). (-16.09 = -17.63 + 1.54)

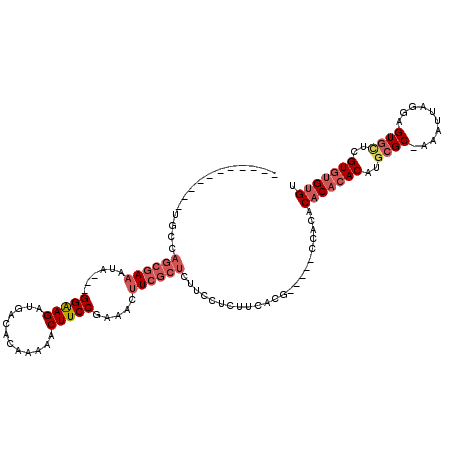

| Location | 8,527,072 – 8,527,172 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.07 |
| Mean single sequence MFE | -33.34 |
| Consensus MFE | -19.12 |
| Energy contribution | -20.47 |
| Covariance contribution | 1.35 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8527072 100 - 22224390 ACACACACGAGCACUCCUAAUUU-GCGCAUGUGUGUGUGUGG-----CGUGAAGAGGAAGAGCGAAGUUUCGGAAGUUUUUGUGCCAUCUUCC---UAUUUCGCUGGCA----------- (((((((((.((.(.........-).)).)))))))))((.(-----.(((((((((((((((((((...(....).))))))....))))))---).))))))).)).----------- ( -36.10) >DroSec_CAF1 1971 100 - 1 ACACUCACGAGCACUCCUAAUUU-GCGCAUGUGUGUGUGUGG-----CGUGAAGAGGAAGAGCGAAGUUUCGGAAGUUUUUAUGUCAUCUUCC---UAUUUCGCUGGCA----------- ((((.((((.((.(.........-).)).)))).))))((.(-----.(((((((((((((..((.....(....)........)).))))))---).))))))).)).----------- ( -29.82) >DroSim_CAF1 1974 100 - 1 ACACACACGAGCACUCCUAAUUU-GCGCAUGUGUGUGUGUGG-----CGUGAAGAGGAAGAGCGAAGUUUCGGAAGUUUUUGUGUCAUCUUCC---UAUUUCGCUGGCA----------- (((((((((.((.(.........-).)).)))))))))((.(-----.(((((((((((((((((((...(....).))))))....))))))---).))))))).)).----------- ( -36.10) >DroEre_CAF1 1913 95 - 1 ---CACACGAGCACUCCUAAUUU-GCGCAUGUGUG--UGUGG-----AGCGAAGAGGAAGAGCGAAGUUUCGGAAGUUUUUGUGUCAUCUUCC---UAUUUCGCUGGCA----------- ---((((((.((.(.........-).)).))))))--(((.(-----.(((((((((((((((((((...(....).))))))....))))))---).))))))).)))----------- ( -32.00) >DroYak_CAF1 1911 96 - 1 --ACACACGAACACUCCUAAUUU-GCGCAUGUGUG--UGUGG-----AGUGUAGAAGAAGAGCGAAGUUUCGGAAGUUUUUGUGUCAUCUUCC---UAUUUCGCUGGAA----------- --........(((((((.....(-((((....)))--)).))-----)))))........((((((((...(((((............)))))---.))))))))....----------- ( -28.40) >DroPer_CAF1 38567 118 - 1 GCACACACAACUACAGUUAAUUUCGCGCAUGUUUGUGUAUGGAUAAGCGCGAAGAGAGGG--AGACGAGACGGAAGCAGCAGUCUCAUCUCCCAUUUUUUUUGUUGGUAUAAUGUAUAAG ....((.((((.........((((((((.(((((......))))).))))))))...(((--(((.(((((..........))))).)))))).........))))))............ ( -37.60) >consensus ACACACACGAGCACUCCUAAUUU_GCGCAUGUGUGUGUGUGG_____CGUGAAGAGGAAGAGCGAAGUUUCGGAAGUUUUUGUGUCAUCUUCC___UAUUUCGCUGGCA___________ (((((((((.((.(..........).)).)))))))))......................((((((((...(((((............)))))....))))))))............... (-19.12 = -20.47 + 1.35)


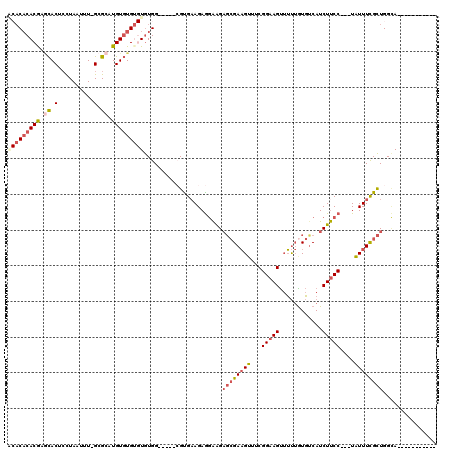
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:05 2006