
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,520,888 – 8,521,073 |
| Length | 185 |
| Max. P | 0.992817 |

| Location | 8,520,888 – 8,521,003 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 93.37 |
| Mean single sequence MFE | -30.74 |
| Consensus MFE | -28.22 |
| Energy contribution | -28.50 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8520888 115 - 22224390 GGGAA-UCCAAAGUGGCCCCCGAAUGUAUGCUGCAAAUUUGCUUAUUAGCU-UACAAAAGGUUUUAAGCAAUUUCGGGGAUUAAUUCACUAAGAACAGGAAGGAUCUUCUCGAAGAU (((((-(((........((((((((((.....)))...(((((((..(.((-(....))).)..))))))).))))))).....(((.((......)))))))))..))))...... ( -30.30) >DroSec_CAF1 62261 116 - 1 GGGAA-UCCAAAGUGGCCCCCGAAUGUAUGCUGCAAAUUUACUUAUUAGCUACACAAAAGUUUUUAAGCAGUUUCGGGGAUUAAUUCACUAGGAACAGGAAGGAUCUUCUCGAAGGU (((((-(((........(((((((.....(((((......((((.............))))......)))))))))))).....(((.((......)))))))))..))))...... ( -26.72) >DroSim_CAF1 65344 116 - 1 GGGAA-UCCAAAGUGGCCCCCGAAUGUAUGCUGCAAAUUUGCUUAUUAGCUAUACAAAAGUUUUUAAGCAAUUUCGGGGAUUAAUUCACUAGGAACAGGAAGGAUCUUCUCGAAGGU (((((-(((........((((((((((.....)))...(((((((..((((.......))))..))))))).))))))).....(((.((......)))))))))..))))...... ( -31.20) >DroEre_CAF1 68581 115 - 1 GGGAU-UCCAUAGUGGCCCCCGAGUGUAUGCAGCAAAUUUGCUUAUUAGCU-UACAAAAGUUUUUAAGCAAUUUCGGGGAUUAAUUCACUAAGAACAGGAAGGAUCUUCUCGGAAGU ....(-(((.((((((.((((((((((.....)))...(((((((..((((-(....)))))..))))))).)))))))......))))))......(((((...))))).)))).. ( -32.90) >DroYak_CAF1 70832 116 - 1 GGGUUUUCCAGUGUGGCCCCCGAAUGUAUGCAGCAAAUUUGCUUAUUAGCU-UACAAAAGUUUUUAAGCAAUUUCGGGGAUUAAUUCACUAAGAACAGGAAGGAUCUUCUCGGAGGU ....(((((((((((..((((((((((.....)))...(((((((..((((-(....)))))..))))))).)))))))..))...)))).......(((((...))))).))))). ( -32.60) >consensus GGGAA_UCCAAAGUGGCCCCCGAAUGUAUGCUGCAAAUUUGCUUAUUAGCU_UACAAAAGUUUUUAAGCAAUUUCGGGGAUUAAUUCACUAAGAACAGGAAGGAUCUUCUCGAAGGU ((((..(((........((((((((((.....)))...(((((((..((((.......))))..))))))).))))))).....(((.((......))))))))...))))...... (-28.22 = -28.50 + 0.28)



| Location | 8,520,928 – 8,521,033 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 87.99 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -23.90 |
| Energy contribution | -23.82 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8520928 105 - 22224390 CCCGAAAAACGCCCGGAUA---------UCCCCCUAUUCGGGAA-UCCAAAGUGGCCCCCGAAUGUAUGCUGCAAAUUUGCUUAUUAGCU-UACAAAAGGUUUUAAGCAAUUUCGG .((((((...((..((...---------.))...((((((((..-.((.....))..))))))))......))....(((((((..(.((-(....))).)..))))))))))))) ( -29.60) >DroSec_CAF1 62301 105 - 1 CCCGAAAAACGCCCGGAUA---------UCCCCC-AUUCGGGAA-UCCAAAGUGGCCCCCGAAUGUAUGCUGCAAAUUUACUUAUUAGCUACACAAAAGUUUUUAAGCAGUUUCGG .((((((...((..((...---------.))..(-(((((((..-.((.....))..))))))))......)).......((((..((((.......))))..))))...)))))) ( -26.40) >DroSim_CAF1 65384 105 - 1 CCCGAAAAACGGCCGGAUA---------UCCCCC-AUUUGGGAA-UCCAAAGUGGCCCCCGAAUGUAUGCUGCAAAUUUGCUUAUUAGCUAUACAAAAGUUUUUAAGCAAUUUCGG .((((((..((((.((...---------.))..(-(((((((..-.((.....))..))))))))...)))).....(((((((..((((.......))))..))))))))))))) ( -32.60) >DroEre_CAF1 68621 104 - 1 CCCGAAAAACCCCCGAAAA---------UCUCCC-AUUCGGGAU-UCCAUAGUGGCCCCCGAGUGUAUGCAGCAAAUUUGCUUAUUAGCU-UACAAAAGUUUUUAAGCAAUUUCGG ............((((((.---------.....(-(((((((..-.((.....))..))))))))............(((((((..((((-(....)))))..))))))))))))) ( -31.00) >DroYak_CAF1 70872 114 - 1 CCCGAAAAACCCCCGGAAAAUAUUCUAUUCUACC-AUUCGGGUUUUCCAGUGUGGCCCCCGAAUGUAUGCAGCAAAUUUGCUUAUUAGCU-UACAAAAGUUUUUAAGCAAUUUCGG .((((((.......((((....)))).......(-(((((((....((.....))..))))))))............(((((((..((((-(....)))))..))))))))))))) ( -30.50) >consensus CCCGAAAAACGCCCGGAUA_________UCCCCC_AUUCGGGAA_UCCAAAGUGGCCCCCGAAUGUAUGCUGCAAAUUUGCUUAUUAGCU_UACAAAAGUUUUUAAGCAAUUUCGG .((((((............................(((((((....((.....))..))))))).............(((((((..((((.......))))..))))))))))))) (-23.90 = -23.82 + -0.08)

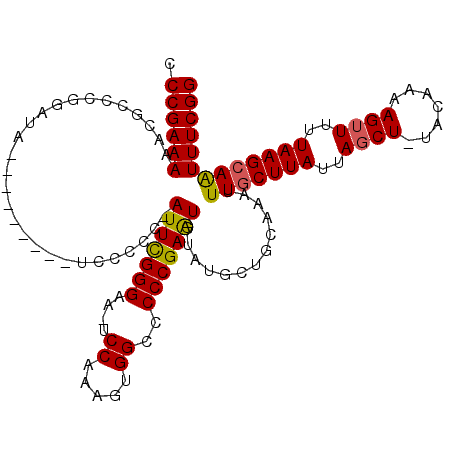

| Location | 8,520,967 – 8,521,073 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 88.39 |
| Mean single sequence MFE | -32.44 |
| Consensus MFE | -22.98 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.790493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8520967 106 + 22224390 UUUGCAGCAUACAUUCGGGGGCCACUUUGGA-UUCCCGAAUAGGGGGA---------UAUCCGGGCGUUUUUCGGGUGAAUCGAAAUAAUUACACAAAGAUGCAGGAAUAAAUGAU ((((((.(....(((((((((((.....)).-)))))))))...((..---------((((((((.....))))))))..))................).)))))).......... ( -27.20) >DroSec_CAF1 62341 105 + 1 UUUGCAGCAUACAUUCGGGGGCCACUUUGGA-UUCCCGAAU-GGGGGA---------UAUCCGGGCGUUUUUCGGGUGAAUCGAAAUAAUUACAGAAAGAUGCUGGAAUAAAUGAU .((.((((((.((((((((((((.....)).-)))))))))-).((..---------((((((((.....))))))))..)).................)))))).))........ ( -34.40) >DroSim_CAF1 65424 105 + 1 UUUGCAGCAUACAUUCGGGGGCCACUUUGGA-UUCCCAAAU-GGGGGA---------UAUCCGGCCGUUUUUCGGGUGAAUCGAAAUAAUUACAGAAAGAUGCUGGAAUAAAUGAU .((.((((((.(.(((...((((.....(((-(((((....-..))))---------.))))))))...((((((.....))))))........))).))))))).))........ ( -31.40) >DroEre_CAF1 68660 105 + 1 UUUGCUGCAUACACUCGGGGGCCACUAUGGA-AUCCCGAAU-GGGAGA---------UUUUCGGGGGUUUUUCGGGUGAACCGAAAUAAUUACAGAAAGAUGCAGGAAUAAAUGAU .((.((((((.((.((((((.((.....)).-.)))))).)-).....---------(((((.(..(((((((((.....)))))).)))..).))))))))))).))........ ( -33.10) >DroYak_CAF1 70911 115 + 1 UUUGCUGCAUACAUUCGGGGGCCACACUGGAAAACCCGAAU-GGUAGAAUAGAAUAUUUUCCGGGGGUUUUUCGGGUGAAUCCAAAUAAUUACAGAAAGAUGCAGGAAUAAAUGAU .((.((((((.(.(((.(((..(((.(((((((((((...(-((.((((((...))))))))).))))))))))))))..)))...........))).))))))).))........ ( -36.10) >consensus UUUGCAGCAUACAUUCGGGGGCCACUUUGGA_UUCCCGAAU_GGGGGA_________UAUCCGGGCGUUUUUCGGGUGAAUCGAAAUAAUUACAGAAAGAUGCAGGAAUAAAUGAU .((.((((((.(((((((((.((.....))...)))))))).)..............((((((((.....)))))))).....................)))))).))........ (-22.98 = -23.90 + 0.92)



| Location | 8,520,967 – 8,521,073 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 88.39 |
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -18.02 |
| Energy contribution | -18.18 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8520967 106 - 22224390 AUCAUUUAUUCCUGCAUCUUUGUGUAAUUAUUUCGAUUCACCCGAAAAACGCCCGGAUA---------UCCCCCUAUUCGGGAA-UCCAAAGUGGCCCCCGAAUGUAUGCUGCAAA ..(((.(((((..((..(((((..(.....(((((.......)))))....((((((((---------......)))))))).)-..)))))..))....))))).)))....... ( -22.50) >DroSec_CAF1 62341 105 - 1 AUCAUUUAUUCCAGCAUCUUUCUGUAAUUAUUUCGAUUCACCCGAAAAACGCCCGGAUA---------UCCCCC-AUUCGGGAA-UCCAAAGUGGCCCCCGAAUGUAUGCUGCAAA ...........((((((...((((......(((((.......)))))......))))..---------.....(-(((((((..-.((.....))..)))))))).)))))).... ( -25.30) >DroSim_CAF1 65424 105 - 1 AUCAUUUAUUCCAGCAUCUUUCUGUAAUUAUUUCGAUUCACCCGAAAAACGGCCGGAUA---------UCCCCC-AUUUGGGAA-UCCAAAGUGGCCCCCGAAUGUAUGCUGCAAA ...........(((((((.(((........(((((.......)))))...((((((((.---------((((..-....)))))-))).....))))...))).).)))))).... ( -27.40) >DroEre_CAF1 68660 105 - 1 AUCAUUUAUUCCUGCAUCUUUCUGUAAUUAUUUCGGUUCACCCGAAAAACCCCCGAAAA---------UCUCCC-AUUCGGGAU-UCCAUAGUGGCCCCCGAGUGUAUGCAGCAAA ...........((((((.............((((((.....))))))............---------.....(-(((((((..-.((.....))..)))))))).)))))).... ( -26.20) >DroYak_CAF1 70911 115 - 1 AUCAUUUAUUCCUGCAUCUUUCUGUAAUUAUUUGGAUUCACCCGAAAAACCCCCGGAAAAUAUUCUAUUCUACC-AUUCGGGUUUUCCAGUGUGGCCCCCGAAUGUAUGCAGCAAA ...........((((((.((((((......(((((......))))).......))))))..............(-(((((((....((.....))..)))))))).)))))).... ( -26.72) >consensus AUCAUUUAUUCCUGCAUCUUUCUGUAAUUAUUUCGAUUCACCCGAAAAACGCCCGGAUA_________UCCCCC_AUUCGGGAA_UCCAAAGUGGCCCCCGAAUGUAUGCUGCAAA ...........(((((((..((((......(((((.......)))))......))))..................(((((((....((.....))..)))))))).)))))).... (-18.02 = -18.18 + 0.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:01 2006