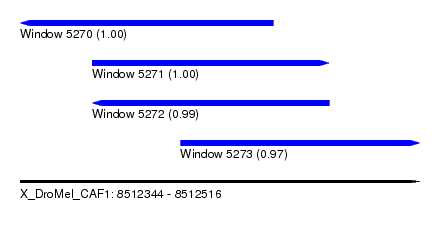
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,512,344 – 8,512,516 |
| Length | 172 |
| Max. P | 0.998088 |

| Location | 8,512,344 – 8,512,453 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.15 |
| Mean single sequence MFE | -37.66 |
| Consensus MFE | -18.18 |
| Energy contribution | -19.10 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8512344 109 - 22224390 CGAGGAAAAGGGGGCUUUUCGGUAGCCCGCCACUUGUAGGCGCGCCACUGAUUAACCCGCAACUCUGCCCCUUUU-CCC----ACUCUUUUCCCCCACCUCCAC-G-----UAGCCCAUU .(.(((((((((((.(..(((((.((.((((.......)))).)).)))))..).)))(((....))).))))))-)))----.....................-.-----......... ( -34.80) >DroSec_CAF1 53529 100 - 1 --------AGGGGGCUUUUCGGUAGCCCGCCACUUGUAGGCGCGGCACUGAUUGACCCGCAACUCUACCCCUUUU-CCC----ACUCUCUUCCCCCAC-UCCCC-G-----UUGCCCAUA --------.(((((.(..(((((.(((((((.......)))).))))))))..).)))(((((............-...----...............-.....-)-----))))))... ( -29.15) >DroSim_CAF1 56734 101 - 1 --------AGGGGGCUUUUCGGUAGCCCGCCACUUGUAGGCGCGGCACUGAUUGACCCGCAACUCUGCCCCUUUUGCCC----CCUCUUCUCCCCCAC-UCCCC-G-----CAGCCCAUA --------(((((((...(((((.(((((((.......)))).)))))))).......(((....))).......))))----)))............-.....-.-----......... ( -34.00) >DroEre_CAF1 60074 109 - 1 UGAGGAAAAGGGGGCUUUUCGGUAGCCCGCCACUUGUAGGCGCGGCACUGAUUGACCCGCAACCCUGCCCCCUUU-UCC----ACUCUUUUCUCCCAC-UCCCCGG-----AAUCCCAUA ...((((((((((((...(((((.(((((((.......)))).))))))))(((.....)))....)))))))))-)))----.........(((...-.....))-----)........ ( -43.60) >DroYak_CAF1 61680 100 - 1 UGAGGAAAAGGGGGCUUUUCGGUAGCCCGCCACUUGUAGGCGCGGCACUGAUUGACCCGCAACUCUGCC-CCUUU-UCC----ACU-------UCCAC-UCCCC-G-----AAUCCCAUA ...(((((((((((.(..(((((.(((((((.......)))).))))))))..).)))(((....))).-)))))-)))----...-------.....-.....-.-----......... ( -37.90) >DroAna_CAF1 86799 117 - 1 CGAUGGCAGGUGGGGUGGAGGGGGGGCGAUCAUUUGUAGGCGCGGCACCGACUGACCCCAAGCCCAAAACCUUCG-CUCCCCAAAGCCCCUCCGCCUC-UUUUC-GAUUUCUUGCCCAAA (((.(((.((.(((((....(((((((((...((((..(((..((((.....)).))....)))))))....)))-))))))...))))).)))))..-...))-).............. ( -46.50) >consensus _GAGGAAAAGGGGGCUUUUCGGUAGCCCGCCACUUGUAGGCGCGGCACUGAUUGACCCGCAACUCUGCCCCUUUU_CCC____ACUCUUUUCCCCCAC_UCCCC_G_____UAGCCCAUA .........(.(((.(..(((((.(((((((.......)))).))))))))..).))).)............................................................ (-18.18 = -19.10 + 0.92)



| Location | 8,512,375 – 8,512,477 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 63.65 |
| Mean single sequence MFE | -39.88 |
| Consensus MFE | -20.64 |
| Energy contribution | -22.57 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.52 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8512375 102 + 22224390 -GGGAAAAGGGGCAGAGUUGCGGGUUAAUCAGUGGCGCGCCUACAAGUGGCGGGCUACCGAAAAGCCCCCUUUUCCUCGCCCCCCCCCCUCCAA-----------AUC-----CCUUUU -(((....(((((.(((..(.(((((..((.(((((.((((.......)))).))))).))..))))).)..)))...)))))...))).....-----------...-----...... ( -42.60) >DroEre_CAF1 60105 96 + 1 -GGAAAAGGGGGCAGGGUUGCGGGUCAAUCAGUGCCGCGCCUACAAGUGGCGGGCUACCGAAAAGCCCCCUUUUCCUCACCCGCCCC----------------------CUUUUCUUUU -((((((((((((.((((.(.(((.........(((((........)))))(((((.......))))).....))).))))))))))----------------------)))))))... ( -48.40) >DroYak_CAF1 61703 105 + 1 -GGAAAAGG-GGCAGAGUUGCGGGUCAAUCAGUGCCGCGCCUACAAGUGGCGGGCUACCGAAAAGCCCCCUUUUCCUCACCCAACCCCA-CCCC-----------AUCCCUCCUUCAUU -((((((((-.((......))((((...((.(((((.((((.......))))))).)).))...)))))))))))).............-....-----------.............. ( -34.20) >DroAna_CAF1 86837 117 + 1 GGAGCGAAGGUUUUGGGCUUGGGGUCAGUCGGUGCCGCGCCUACAAAUGAUCGCCCCCCCUCCACCCCACCUGCCAUCGCACUUCUCCA-CCCCUUGGAGGCCCUACCCCCACCCCCU- ((.((((.(((..((((...(((((((((.((((...)))).))...)))......)))).....))))...))).))))...((((((-.....))))))........)).......- ( -34.30) >consensus _GGAAAAAGGGGCAGAGUUGCGGGUCAAUCAGUGCCGCGCCUACAAGUGGCGGGCUACCGAAAAGCCCCCUUUUCCUCACCCACCCCCA_CCCC___________AUC_CU_CCCCUUU .((((((((((((.......((((.....)...(((.((((.......)))))))..)))....))))))))))))........................................... (-20.64 = -22.57 + 1.94)

| Location | 8,512,375 – 8,512,477 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 63.65 |
| Mean single sequence MFE | -47.90 |
| Consensus MFE | -21.60 |
| Energy contribution | -22.35 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.45 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8512375 102 - 22224390 AAAAGG-----GAU-----------UUGGAGGGGGGGGGGCGAGGAAAAGGGGGCUUUUCGGUAGCCCGCCACUUGUAGGCGCGCCACUGAUUAACCCGCAACUCUGCCCCUUUUCCC- ......-----...-----------.....((..((((((((((.....(.(((.(..(((((.((.((((.......)))).)).)))))..).))).)..))).)))))))..)).- ( -45.20) >DroEre_CAF1 60105 96 - 1 AAAAGAAAAG----------------------GGGGCGGGUGAGGAAAAGGGGGCUUUUCGGUAGCCCGCCACUUGUAGGCGCGGCACUGAUUGACCCGCAACCCUGCCCCCUUUUCC- ....((((((----------------------((((((((.(..(....(((..(...(((((.(((((((.......)))).))))))))..).))).)..))))))))))))))).- ( -51.90) >DroYak_CAF1 61703 105 - 1 AAUGAAGGAGGGAU-----------GGGG-UGGGGUUGGGUGAGGAAAAGGGGGCUUUUCGGUAGCCCGCCACUUGUAGGCGCGGCACUGAUUGACCCGCAACUCUGCC-CCUUUUCC- .........((((.-----------((((-..(((((((((.((((((((....))))))(((.(((((((.......)))).))))))..)).))))..)))))..))-))..))))- ( -46.50) >DroAna_CAF1 86837 117 - 1 -AGGGGGUGGGGGUAGGGCCUCCAAGGGG-UGGAGAAGUGCGAUGGCAGGUGGGGUGGAGGGGGGGCGAUCAUUUGUAGGCGCGGCACCGACUGACCCCAAGCCCAAAACCUUCGCUCC -..(((((((((((.(((((((((.....-)))))...(((....)))..((((((.(.(..((.(((.((.......)))))....))..)).)))))).))))...))))))))))) ( -48.00) >consensus AAAAGAG_AG_GAU___________GGGG_UGGGGGAGGGCGAGGAAAAGGGGGCUUUUCGGUAGCCCGCCACUUGUAGGCGCGGCACUGAUUGACCCGCAACCCUGCCCCCUUUCCC_ ................................(((..(((...(((...(.(((....(((((.(((((((.......)))).))))))))....))).)...)))....)))..))). (-21.60 = -22.35 + 0.75)



| Location | 8,512,413 – 8,512,516 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 83.85 |
| Mean single sequence MFE | -20.57 |
| Consensus MFE | -17.80 |
| Energy contribution | -17.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8512413 103 + 22224390 CCUACAAGUGGCGGGCUACCGAAAAGCCCCCUUUUCCUCGCCCCCCCCCCUCCAAAUC-----CCUUUUCUUUCCUAAUUUGACCCAGCUUAUCCAAGGCGUUGUCAU .........((.((((....((((((....))))))...)))))).............-----.................((((...((((.....))))...)))). ( -20.30) >DroEre_CAF1 60143 97 + 1 CCUACAAGUGGCGGGCUACCGAAAAGCCCCCUUUUCCUCACCCGCCCC-----------CUUUUCUUUUCUUUCCUAAUUUGACCCAGCUUAUCCAAGGCGUUGUCAU .........((((((.....((((((....))))))....))))))..-----------.....................((((...((((.....))))...)))). ( -23.00) >DroYak_CAF1 61740 107 + 1 CCUACAAGUGGCGGGCUACCGAAAAGCCCCCUUUUCCUCACCCAACCCCA-CCCCAUCCCUCCUUCAUUCGUUCCUAAUUUGACCCAGCUUAUCCAAGGCGUUGUCAU .......((((.(((.....((((((....))))))....)))....)))-)............................((((...((((.....))))...)))). ( -18.40) >consensus CCUACAAGUGGCGGGCUACCGAAAAGCCCCCUUUUCCUCACCCACCCCC__CC__AUC_CU__UCUUUUCUUUCCUAAUUUGACCCAGCUUAUCCAAGGCGUUGUCAU .........((.(((((.......))))))).................................................((((...((((.....))))...)))). (-17.80 = -17.80 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:57 2006