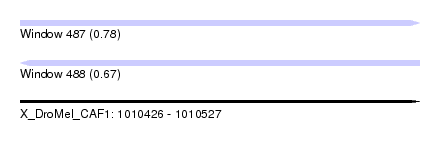
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,010,426 – 1,010,527 |
| Length | 101 |
| Max. P | 0.775602 |

| Location | 1,010,426 – 1,010,527 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.04 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -23.94 |
| Energy contribution | -24.50 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
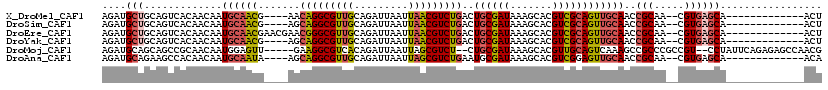
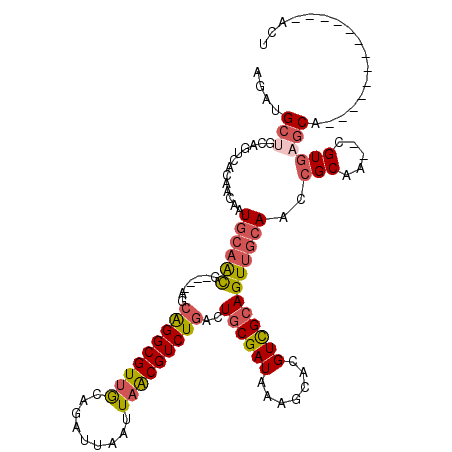
>X_DroMel_CAF1 1010426 101 + 22224390 AGU-------------UGCUCACG--UUGCGGUUGCAACUGCGACGUGCUUUAUCGCAGUCAGACGUUAAUUAAUCUGCAACGCCUGUU----CGUUGCAUUGUUGUGACUGCAGCAUCU ((.-------------((((((((--(((((((....))))))))))).......(((((((((((..........(((((((......----))))))).)))).))))))))))).)) ( -39.30) >DroSim_CAF1 73094 101 + 1 AGU-------------UGCUCACG--UUGCGGUUGCAACUGCGACGUGCUUUAUCGCAGUCAGACGUUAAUUAAUCUGCAACGCCUGCU----CGUUGCAUUGUUGUGACUGCAGCAUCU ((.-------------((((((((--(((((((....))))))))))).......(((((((((((..........(((((((......----))))))).)))).))))))))))).)) ( -39.30) >DroEre_CAF1 107260 105 + 1 AGU-------------UGCUCACG--UUGCGGUUGCAACUGCGACGUGCUUUAUCGCAGUCAGACGUUAAUUAAUCUGCAACGCCCGUUCGUUCGUUGCAUUGUUGUGACUGCAGCAUCU ((.-------------((((((((--(((((((....))))))))))).......(((((((((((..........(((((((..(....)..))))))).)))).))))))))))).)) ( -40.60) >DroYak_CAF1 98761 101 + 1 AGU-------------UGCUCACG--UUGCGGUUGCAACUGCGACGUGCUUUAUCGCAGUCAGACGUUAAUUAAUCUGCAACGCCUGCU----CGUUGCAUUGUUGUGACUGCAGCAUCU ((.-------------((((((((--(((((((....))))))))))).......(((((((((((..........(((((((......----))))))).)))).))))))))))).)) ( -39.30) >DroMoj_CAF1 185866 111 + 1 CGUUGGCUCUCUGAAUAGG--ACGGCGGGCGGCUUUGACUGCAACGUGCUUUAUCGCAG--AGACGCUAAUUAAUCUGUGACGCCUUC-----AACUCCAUUGUUGCGGCUGCUGCAUCU .((((((((((((...((.--(((....((((......))))..))).))......)))--))).)))))).....(((..((((..(-----(((......)))).))).)..)))... ( -34.20) >DroAna_CAF1 98603 101 + 1 UGU-------------UGCUCACG--UUGCGGUUGCAACUCCGACGUGCUUUAUCGCAUUCAGACGCUAAUUAAUCUGCAACGCCUGCU----UAUUGCAUUGUUGUGGCUUCUGCAUCU ...-------------(((.((((--(((.(((....))).))))))).............(((.(((.........((((((..(((.----....))).))))))))).))))))... ( -23.30) >consensus AGU_____________UGCUCACG__UUGCGGUUGCAACUGCGACGUGCUUUAUCGCAGUCAGACGUUAAUUAAUCUGCAACGCCUGCU____CGUUGCAUUGUUGUGACUGCAGCAUCU ...........................((((((..((((...(((.(((......))))))...............(((((((..........)))))))..))))..))))))...... (-23.94 = -24.50 + 0.56)

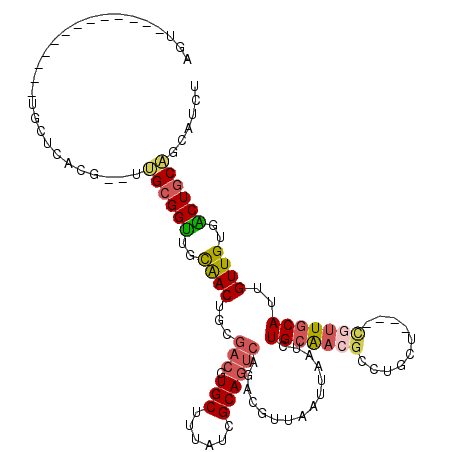
| Location | 1,010,426 – 1,010,527 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.04 |
| Mean single sequence MFE | -31.87 |
| Consensus MFE | -21.40 |
| Energy contribution | -22.07 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1010426 101 - 22224390 AGAUGCUGCAGUCACAACAAUGCAACG----AACAGGCGUUGCAGAUUAAUUAACGUCUGACUGCGAUAAAGCACGUCGCAGUUGCAACCGCAA--CGUGAGCA-------------ACU ...(((((((((((......(((((((----......)))))))(((........)))))))))).......(((((.((.((....)).)).)--))))))))-------------... ( -34.00) >DroSim_CAF1 73094 101 - 1 AGAUGCUGCAGUCACAACAAUGCAACG----AGCAGGCGUUGCAGAUUAAUUAACGUCUGACUGCGAUAAAGCACGUCGCAGUUGCAACCGCAA--CGUGAGCA-------------ACU ...(((((((((((......(((((((----......)))))))(((........)))))))))).......(((((.((.((....)).)).)--))))))))-------------... ( -34.00) >DroEre_CAF1 107260 105 - 1 AGAUGCUGCAGUCACAACAAUGCAACGAACGAACGGGCGUUGCAGAUUAAUUAACGUCUGACUGCGAUAAAGCACGUCGCAGUUGCAACCGCAA--CGUGAGCA-------------ACU ...(((((((((((......(((((((..(....)..)))))))(((........)))))))))).......(((((.((.((....)).)).)--))))))))-------------... ( -36.40) >DroYak_CAF1 98761 101 - 1 AGAUGCUGCAGUCACAACAAUGCAACG----AGCAGGCGUUGCAGAUUAAUUAACGUCUGACUGCGAUAAAGCACGUCGCAGUUGCAACCGCAA--CGUGAGCA-------------ACU ...(((((((((((......(((((((----......)))))))(((........)))))))))).......(((((.((.((....)).)).)--))))))))-------------... ( -34.00) >DroMoj_CAF1 185866 111 - 1 AGAUGCAGCAGCCGCAACAAUGGAGUU-----GAAGGCGUCACAGAUUAAUUAGCGUCU--CUGCGAUAAAGCACGUUGCAGUCAAAGCCGCCCGCCGU--CCUAUUCAGAGAGCCAACG .(((((..((((..((....))..)))-----)...)))))............((.(((--(((.((((..((.((..((.((....)).)).))..))--..))))))))))))..... ( -25.60) >DroAna_CAF1 98603 101 - 1 AGAUGCAGAAGCCACAACAAUGCAAUA----AGCAGGCGUUGCAGAUUAAUUAGCGUCUGAAUGCGAUAAAGCACGUCGGAGUUGCAACCGCAA--CGUGAGCA-------------ACA ..........(((.......(((....----.))))))(((((.............(((((.(((......)))..)))))(((((....))))--)....)))-------------)). ( -27.21) >consensus AGAUGCUGCAGUCACAACAAUGCAACG____AGCAGGCGUUGCAGAUUAAUUAACGUCUGACUGCGAUAAAGCACGUCGCAGUUGCAACCGCAA__CGUGAGCA_____________ACU ....(((.............((((((.......(((((((((.........)))))))))..((((((.......))))))))))))..(((.....))))))................. (-21.40 = -22.07 + 0.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:40 2006