
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,487,710 – 8,487,870 |
| Length | 160 |
| Max. P | 0.909405 |

| Location | 8,487,710 – 8,487,830 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.44 |
| Mean single sequence MFE | -55.73 |
| Consensus MFE | -50.17 |
| Energy contribution | -48.98 |
| Covariance contribution | -1.19 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
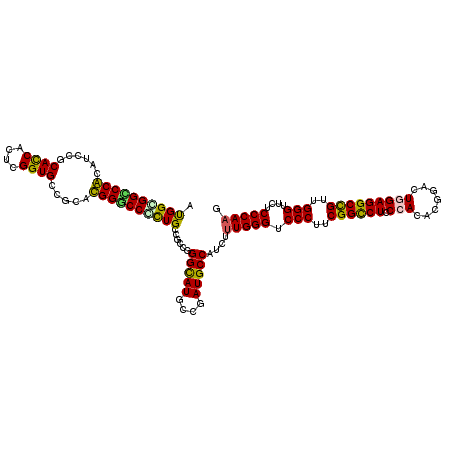
>X_DroMel_CAF1 8487710 120 - 22224390 ACGGGGGUCCGCAUCCGCAUCAUUCGGUGCCGCACGGACCCCUGCCGCCCGGCAUGCCCAUGCCAUCGCUGGGGCCCUUCGGCCUCCCACACGGACUGGAGGCCGUCGGGUUCUCCCAAG .((((((((((...(.(((((....))))).)..))))))))))..((..(((((....)))))...))((((((((..((((((((..........))))))))..))))...)))).. ( -62.90) >DroVir_CAF1 11227 120 - 1 AUGGCGGGCCACAUCCGCACCAUUCGGUGACGCAUGGACCGCUGCCGCCGGGCAUGCCAAUGCCAUCUUUGGGUCCCUUCGGCCUGCCACACGGACUGGAGGCUGUUGGGUUCUCCCAAG .((((((((..(((.((((((....)))).)).)))....))..))))))(((((....)))))....(((((.(((..((((((.(((.......)))))))))..)))....))))). ( -52.90) >DroPse_CAF1 35929 120 - 1 AUGGCGGCCCGCAUCCCCAUCACUCGGUGCCGCACGGGCCCCUGCCGCCGGGCAUGCCGAUGCCGUCUUUGGGUCCCUUCGGCCUCCCACAUGGUCUCGAGGCCGUUGGGUUCUCCCAAG ..((((((..((((.(((.......((.(((.....))).)).......))).))))....)))))).(((((.(((..(((((((((....))....)))))))..)))....))))). ( -53.04) >DroGri_CAF1 11232 120 - 1 AUGGCGGCCCACAUCCGCACCACUCGGUGCCGCAUGGGCCGCUGCCGCCGGGCAUGCCGAUGCCAUCGUUGGGUCCCUUCGGCCUGCCACACGGACUGGAGGCCGUUGGGUUCUCCCAAG (((((((((((...(.(((((....))))).)..))))))((((((....)))).))....)))))..(((((.(((..((((((.(((.......)))))))))..)))....))))). ( -62.10) >DroWil_CAF1 9875 120 - 1 AUGGUGGGCCACAUCCGCACCAUUCGGUGCCUCAUGGCCCAUUGCCGCCGGGUAUGCCGAUGCCGUCAUUGGGUCCCUUCGGUCUGCCACAUGGUCUGGAGGCUGUUGGGUUCUCCCAAG ..(((((((((.....(((((....)))))....)))))))))(((.((((....(((((.(((.......)))....)))))..(((....))))))).)))..(((((....))))). ( -50.40) >DroPer_CAF1 38535 120 - 1 AUGGCGGCCCGCAUCCCCAUCACUCGGUGCCGCACGGGCCCCUGCCGCCGGGCAUGCCGAUGCCGUCUUUGGGUCCCUUCGGCCUCCCACAUGGUCUCGAGGCCGUUGGGUUCUCCCAAG ..((((((..((((.(((.......((.(((.....))).)).......))).))))....)))))).(((((.(((..(((((((((....))....)))))))..)))....))))). ( -53.04) >consensus AUGGCGGCCCACAUCCGCACCACUCGGUGCCGCACGGGCCCCUGCCGCCGGGCAUGCCGAUGCCAUCUUUGGGUCCCUUCGGCCUCCCACACGGACUGGAGGCCGUUGGGUUCUCCCAAG .((((((((((......((((....)))).....))))))))))......(((((....)))))....(((((.(((..((((((.(((.......)))))))))..)))....))))). (-50.17 = -48.98 + -1.19)


| Location | 8,487,750 – 8,487,870 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.33 |
| Mean single sequence MFE | -48.33 |
| Consensus MFE | -39.78 |
| Energy contribution | -38.65 |
| Covariance contribution | -1.13 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8487750 120 - 22224390 UUCCUCAAAUCGGGCGAUUUGGGCCCCCAUCCGCACAGUUACGGGGGUCCGCAUCCGCAUCAUUCGGUGCCGCACGGACCCCUGCCGCCCGGCAUGCCCAUGCCAUCGCUGGGGCCCUUC ......(((((....)))))(((((((.....((...((..((((((((((...(.(((((....))))).)..))))))))))..))..(((((....)))))...)).)))))))... ( -54.50) >DroVir_CAF1 11267 120 - 1 UUUCUCAAAUCGGGUGAUUUGGGCCCACAUCCGCACAGCUAUGGCGGGCCACAUCCGCACCAUUCGGUGACGCAUGGACCGCUGCCGCCGGGCAUGCCAAUGCCAUCUUUGGGUCCCUUC ......(((((....)))))(((((((.....((.((((....(((((.....))))).((((.((....)).))))...))))..))..(((((....))))).....))))))).... ( -44.40) >DroPse_CAF1 35969 120 - 1 UUUCUCAAAUCUGGCGAUUUGGGCCCCCAUCCGCACAGCUAUGGCGGCCCGCAUCCCCAUCACUCGGUGCCGCACGGGCCCCUGCCGCCGGGCAUGCCGAUGCCGUCUUUGGGUCCCUUC ...((((((..(((((.(..((((((.(((..((...)).)))((((((((.............))).)))))..))))))..).)))))(((((....)))))...))))))....... ( -44.82) >DroGri_CAF1 11272 120 - 1 UUUCUCAAAUCAGGUGAUUUGGGCCCACAUCCGCACAGCUAUGGCGGCCCACAUCCGCACCACUCGGUGCCGCAUGGGCCGCUGCCGCCGGGCAUGCCGAUGCCAUCGUUGGGUCCCUUC ......(((((....)))))(((((((((((.(((.......(((((((((...(.(((((....))))).)..)))))))))(((....))).))).)))).......))))))).... ( -51.71) >DroWil_CAF1 9915 120 - 1 UUUCUCAAAUCGGGUGAUUUGGGGCCACAUCCGCAUAGCUAUGGUGGGCCACAUCCGCACCAUUCGGUGCCUCAUGGCCCAUUGCCGCCGGGUAUGCCGAUGCCGUCAUUGGGUCCCUUC ...........(((.((((..((((..((((.(((((.((.((((((((((.....(((((....)))))....))))))...))))..)).))))).))))..))).)..))))))).. ( -48.20) >DroAna_CAF1 50460 120 - 1 UUCCUCAAAUCGGGCGAUUUGGGCCCCCAUCCGCACAGUUACGGGGGCCCGCACCCCCACCACUCGGUGCCGCACGGUCCCCUGCCGCCCGGAAUGCCGAUGCCAUCGCUGGGUCCCUUC ........(((.((((((..((((((((..............))))))))(((..........(((((.(((..((((.....))))..)))...)))))))).)))))).)))...... ( -46.34) >consensus UUUCUCAAAUCGGGCGAUUUGGGCCCACAUCCGCACAGCUAUGGCGGCCCACAUCCGCACCACUCGGUGCCGCACGGACCCCUGCCGCCGGGCAUGCCGAUGCCAUCGUUGGGUCCCUUC ......(((((....)))))((((((...........((..((((((((((......((((....)))).....))))))))))..))..(((((....)))))......)))))).... (-39.78 = -38.65 + -1.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:47 2006