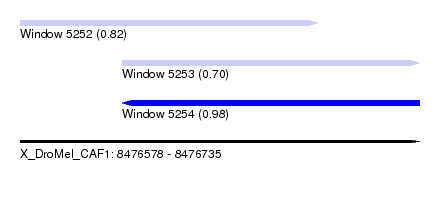
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,476,578 – 8,476,735 |
| Length | 157 |
| Max. P | 0.977367 |

| Location | 8,476,578 – 8,476,695 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 92.98 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -24.08 |
| Energy contribution | -24.57 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8476578 117 + 22224390 UUCAUAAUGUUAUGUCUUAGUUUGGUUUAAAGAACUUGAAAUUGGAAUUAAGUUUUAGCUUUAUGAUAUGCUGAGCUGGUCCAAACCACCAGUUCUAUGGUGGUGGUGGUGUUGGUG .((((((.((((...(((((((..((((.((....)).))))..).))))))...)))).))))))........(((..((((.(((((((......)))))))..))).)..))). ( -33.20) >DroSec_CAF1 19178 111 + 1 UUCGUAAUGUUAUGUCUUAGUUUGGUUUAAAGAACUUGAAAUUUGAAUUAAGUUUUAGCUUUAUGAUAUGCUGAGCUGGUCCGAACCACCAGUUCUAUGGUGAUGGUG------UUG .((((((.((((...(((((((..((((((.....)))))..)..)))))))...)))).))))))...(((((((((((.......))))))))...))).......------... ( -25.70) >DroSim_CAF1 28294 111 + 1 UUCGUAAUGUUAUGUCUUAGUUUGGUUUAAAGAACUUGAAAUUGGAAUUAAGUUUUAGCUUUAUGAUAUGCUGAGCUGGUCCAAACCACCAGUUCUAUGCUGAUGGUG------GUG .((((((.((((...(((((((..((((.((....)).))))..).))))))...)))).))))))...((.((((((((.......))))))))...))........------... ( -27.10) >DroEre_CAF1 24953 111 + 1 UUCGUAAUGUUAUGUCUUAGUUUGGUUUAAAGAACUUGAAAUUGGAAUUAAGUUUUAGCUUUAUGAUAUGCUGAGCUGGUCCAAGCCACCACUUUUAUGGUGGUGGUG------CUG .((((((.((((...(((((((..((((.((....)).))))..).))))))...)))).)))))).......(((........(((((((((.....))))))))))------)). ( -30.30) >consensus UUCGUAAUGUUAUGUCUUAGUUUGGUUUAAAGAACUUGAAAUUGGAAUUAAGUUUUAGCUUUAUGAUAUGCUGAGCUGGUCCAAACCACCAGUUCUAUGGUGAUGGUG______GUG ............((..((((((..((.((((((((((((........)))))))....)))))......))..))))))..)).(((((((((.....))))))))).......... (-24.08 = -24.57 + 0.50)

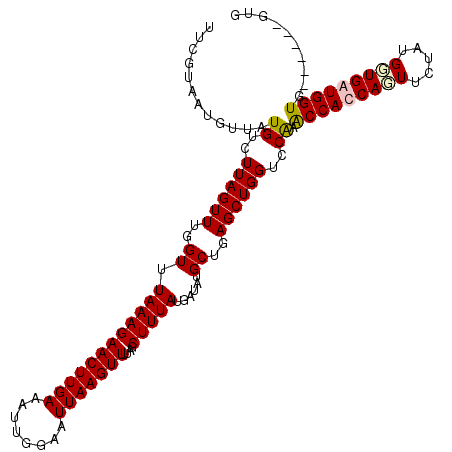

| Location | 8,476,618 – 8,476,735 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 90.01 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -25.83 |
| Energy contribution | -26.57 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8476618 117 + 22224390 AUUGGAAUUAAGUUUUAGCUUUAUGAUAUGCUGAGCUGGUCCAAACCACCAGUUCUAUGGUGGUGGUGGUGUUGGUGGUGCAGCUCUGUUCUCCUCUUACUGGCGAUCCCCAAAUGU .(((((....(((((.(((..........))))))))..)))))(((((((......)))))))((.(((((..((((.(((....)))...))....))..)))..))))...... ( -34.70) >DroSec_CAF1 19218 110 + 1 AUUUGAAUUAAGUUUUAGCUUUAUGAUAUGCUGAGCUGGUCCGAACCACCAGUUCUAUGGUGAUGGUG------UUGGUGCAGCUCUGUUCUCUUAUUUCUGGCGAUCCCCAAAA-C ....(((.((((....(((..........)))((((((..(((((((((((......))))...))).------))))..))))))......)))).)))(((......)))...-. ( -26.60) >DroSim_CAF1 28334 110 + 1 AUUGGAAUUAAGUUUUAGCUUUAUGAUAUGCUGAGCUGGUCCAAACCACCAGUUCUAUGCUGAUGGUG------GUGGUGCAGCUCUGUUCUCUUAUUUCUGGCGAUCCCCAAAA-C .(..(((.((((....(((..........)))((((((..(((.((((.((((.....)))).)))).------.)))..))))))......)))).)))..)............-. ( -31.80) >DroEre_CAF1 24993 110 + 1 AUUGGAAUUAAGUUUUAGCUUUAUGAUAUGCUGAGCUGGUCCAAGCCACCACUUUUAUGGUGGUGGUG------CUGGUGCAGCUCUGUUUUCUUAUUUCUGGCGAUCCCCAAAG-C .(..(((.((((....(((..........)))((((((..(((.(((((((((.....))))))))).------.)))..))))))......)))).)))..)............-. ( -38.20) >consensus AUUGGAAUUAAGUUUUAGCUUUAUGAUAUGCUGAGCUGGUCCAAACCACCAGUUCUAUGGUGAUGGUG______GUGGUGCAGCUCUGUUCUCUUAUUUCUGGCGAUCCCCAAAA_C .................(((....((((.((.((((((..(((.(((((((((.....)))))))))........)))..)))))).)).....))))...)))............. (-25.83 = -26.57 + 0.75)


| Location | 8,476,618 – 8,476,735 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 90.01 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -25.11 |
| Energy contribution | -26.48 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 8476618 117 - 22224390 ACAUUUGGGGAUCGCCAGUAAGAGGAGAACAGAGCUGCACCACCAACACCACCACCACCAUAGAACUGGUGGUUUGGACCAGCUCAGCAUAUCAUAAAGCUAAAACUUAAUUCCAAU ....((((......)))).....((((....((((((.....((((.(((((((............)))))))))))..))))))(((..........))).........))))... ( -29.90) >DroSec_CAF1 19218 110 - 1 G-UUUUGGGGAUCGCCAGAAAUAAGAGAACAGAGCUGCACCAA------CACCAUCACCAUAGAACUGGUGGUUCGGACCAGCUCAGCAUAUCAUAAAGCUAAAACUUAAUUCAAAU .-...(((......)))(((.((((......((((((..((..------.(((((((.........)))))))..))..))))))(((..........)))....)))).))).... ( -26.60) >DroSim_CAF1 28334 110 - 1 G-UUUUGGGGAUCGCCAGAAAUAAGAGAACAGAGCUGCACCAC------CACCAUCAGCAUAGAACUGGUGGUUUGGACCAGCUCAGCAUAUCAUAAAGCUAAAACUUAAUUCCAAU .-((((((......)))))).((((......((((((..(((.------.((((((((.......)))))))).)))..))))))(((..........)))....))))........ ( -31.40) >DroEre_CAF1 24993 110 - 1 G-CUUUGGGGAUCGCCAGAAAUAAGAAAACAGAGCUGCACCAG------CACCACCACCAUAAAAGUGGUGGCUUGGACCAGCUCAGCAUAUCAUAAAGCUAAAACUUAAUUCCAAU .-..((((((.((....))..((((......((((((..((((------..(((((((.......))))))).))))..))))))(((..........)))....)))).)))))). ( -32.60) >consensus G_UUUUGGGGAUCGCCAGAAAUAAGAGAACAGAGCUGCACCAC______CACCACCACCAUAGAACUGGUGGUUUGGACCAGCUCAGCAUAUCAUAAAGCUAAAACUUAAUUCCAAU .....(((......)))(((.((((......((((((..((((.......((((((((.......))))))))))))..))))))(((..........)))....)))).))).... (-25.11 = -26.48 + 1.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:40 2006