| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 8,472,703 – 8,472,802 |
| Length | 99 |
| Max. P | 0.837370 |

| Location | 8,472,703 – 8,472,802 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.24 |
| Mean single sequence MFE | -34.28 |
| Consensus MFE | -14.57 |
| Energy contribution | -16.04 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
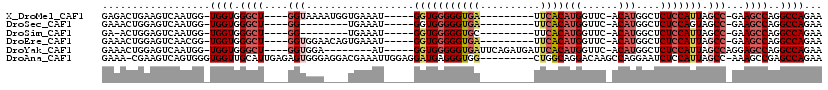
>X_DroMel_CAF1 8472703 99 + 22224390 GAGACUGAAGUCAAUGG-UGGUGGGCU----GGUAAAAUGGUGAAAU-----GGUGGGGGUGA---------UUCACAUGGUUC-ACAUGGCUCUCCAUUAGCC-GAAGCCAGGCCAGAA ..(((....))).....-...(((.((----(((....((((..(((-----((.(((.((((---------..(....)..))-))....))).))))).)))-)..))))).)))... ( -32.80) >DroSec_CAF1 15370 91 + 1 GAAACUGGAGUCAAUGG-UGGUGGGCU----GG--------UGAAAU-----GGUGGGGGUGA---------UUCACAUGGUUC-ACAUGGCUCUCCAGUAGCC-GAAGCCAGGCCAGAA ....((((..((..((.-(((.(((((----((--------((((..-----.(((((.....---------)))))....)))-)).)))))).))).))...-))..))))....... ( -32.50) >DroSim_CAF1 24469 90 + 1 GA-ACUGGAGUCAAUGG-UGGUGGGCU----GG--------UGAAAU-----GGUGGGGGUGC---------UUCACAUGGUUC-ACAUGGCUCUCCAUUAGCC-GAAGCCAGGCCAGAA (.-.((((..((..(((-(((.(((((----((--------((((..-----.((((((...)---------)))))....)))-)).)))))).))))))...-))..))))..).... ( -35.80) >DroEre_CAF1 21507 99 + 1 GAAACUGGAGUCAACGG-UGGUGGGCU----GGUGGAACAGUGAAAU-----GGUGGGGGUGA---------UUCACAUGGUUC-ACAUGGCUCUCCAUUAGCC-GAAGCCAGGCCAGAA ....((((.(....)((-(..(.((((----((((((.(((((((..-----.(((((.....---------)))))....)))-)).))....))))))))))-.).)))...)))).. ( -35.90) >DroYak_CAF1 22227 101 + 1 GAAACUGGAGUCAAUGG-UGGUGGGCU----GGUGGA--------AU-----GGUGGGGGUGAUUCAGAUGAUUCACAUGGUUC-ACAUGGCUCUCCAUUAGCCAGGAGCCAGGCCAGAA ....((((......(((-(....((((----((((((--------..-----(((.(..((((..((..((...))..))..))-)).).))).))))))))))....))))..)))).. ( -36.80) >DroAna_CAF1 31968 109 + 1 GAAA-CGAAGUCAGUGGGUGGUUGCAUUGAGAGUGGGAGGACGAAAUUGGAGGAUGAGGGUGG---------CUGGCAGGACAAGCCAGGAAUCUCCAUUAGCC-AAAGCCGAGCCAGAA ....-((...((((((.(....).))))))...))((.((.(....((((..((((..(((..---------(((((.......)))))..)))..))))..))-)).)))...)).... ( -31.90) >consensus GAAACUGGAGUCAAUGG_UGGUGGGCU____GGUGGA____UGAAAU_____GGUGGGGGUGA_________UUCACAUGGUUC_ACAUGGCUCUCCAUUAGCC_GAAGCCAGGCCAGAA ..................((((.((((....(((..................(((((((((((..........))))(((......)))....))))))).)))...))))..))))... (-14.57 = -16.04 + 1.47)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:38 2006